kmers
$ lexicmap utils kmers -h
View k-mers captured by the masks
Attention:
1. Mask index (column mask) is 1-based.
2. Prefix means the length of shared prefix between a k-mer and the mask.
3. K-mer positions (column pos) are 1-based.
For reference genomes with multiple sequences, the sequences were
concatenated to a single sequence with intervals of N's.
4. Reversed means if the k-mer is reversed for suffix matching.
Usage:
lexicmap utils kmers [flags] -d <index path> [-m <mask index>] [-o out.tsv.gz]
Flags:
-h, --help help for kmers
-d, --index string ► Index directory created by "lexicmap index".
-m, --mask int ► View k-mers captured by Xth mask. (0 for all) (default 1)
-f, --only-forward ► Only output forward k-mers.
-o, --out-file string ► Out file, supports and recommends a ".gz" suffix ("-" for stdout).
(default "-")
Global Flags:
-X, --infile-list string ► File of input file list (one file per line). If given, they are
appended to files from CLI arguments.
--log string ► Log file.
--quiet ► Do not print any verbose information. But you can write them to a file
with --log.
-j, --threads int ► Number of CPU cores to use. By default, it uses all available cores.
(default 16)
-
The default output is captured k-mers of the first mask.
$ lexicmap utils kmers --quiet -d demo.lmi/ | head -n 20 | csvtk pretty -t mask kmer prefix number ref pos strand reversed ---- ------------------------------- ------ ------ --------------- ------- ------ -------- 1 AAAAAAAAACGAAAAAGATTTTCCCTCATAC 7 1 GCF_000392875.1 2088530 + yes 1 AAAAAAAAACGCTTCTACATCGAGCAGCGAG 7 1 GCF_001457655.1 941619 + yes 1 AAAAAAAAACGTATCCCTCTTTATTACTTAT 7 1 GCF_000006945.2 3392260 - yes 1 AAAAAAAAAGATTTGATTTTTTTCATTAATA 7 1 GCF_000392875.1 766998 - yes 1 AAAAAAAAATCTATTTTAAAACCTAATCACG 7 1 GCF_000392875.1 2201506 + yes 1 AAAAAAAAATGTCACAACAGCCCAACCTCCA 7 1 GCF_000392875.1 860216 + yes 1 AAAAAAAACAAAAACTAGTTCGAGTGCCGAA 7 1 GCF_000006945.2 1587885 - yes 1 AAAAAAAACCATATTATGTCCGATCCTCACA 7 1 GCF_000392875.1 1060650 + yes 1 AAAAAAAACGAAAAACGGTAACACGGGAATT 7 1 GCF_001544255.1 1605298 + yes 1 AAAAAAAACGACGCAGAAAACGACATTGCGA 7 1 GCF_003697165.2 564733 + yes 1 AAAAAAAACGACTCCAGAGAGATCATCGTAT 7 1 GCF_000392875.1 1279686 + yes 1 AAAAAAAACGAGCGATTGGTTGCATTAAGGA 7 1 GCF_002949675.1 3914985 - yes 1 AAAAAAAACGAGCGCTCGGTTGCATTAAGGA 7 2 GCF_002949675.1 2061956 - yes 1 AAAAAAAACGAGCGCTCGGTTGCATTAAGGA 7 2 GCF_003697165.2 1514669 - yes 1 AAAAAAAACGCAACTTAAACAGTAAAACACG 7 1 GCF_002950215.1 1938205 + yes 1 AAAAAAAACGGGACGCGTAGTGCTGTGGTCT 7 1 GCF_000742135.1 2728620 - yes 1 AAAAAAAACGTAAATTTTTAAGATTGCGTCG 7 1 GCF_001457655.1 1547239 - yes 1 AAAAAAAACGTTAGAGAAAGCATCTAACACA 7 1 GCF_001027105.1 660296 + yes 1 AAAAAAAACGTTTTATCACTAATTTTCAGTT 7 1 GCF_000392875.1 1590621 - yesOnly forward k-mers.
$ lexicmap utils kmers --quiet -d demo.lmi/ -f | head -n 20 | csvtk pretty -t mask kmer prefix number ref pos strand reversed ---- ------------------------------- ------ ------ --------------- ------- ------ -------- 1 AAAAAAATAAAAACTTAGTTGTCCCATAACA 8 1 GCF_000392875.1 1044207 - no 1 AAAAAAATAAATCTGCGATGGCTGTTGATGG 8 1 GCF_002950215.1 462416 + no 1 AAAAAAATAACGTTGGCGATTACGATGCCAA 8 1 GCF_000392875.1 1422018 + no 1 AAAAAAATAACTCAATGAGGTTATGGGCATG 8 1 GCF_000742135.1 4160317 - no 1 AAAAAAATAACTGCTTTACTCTTTGCTCTTT 8 1 GCF_009759685.1 2134145 + no 1 AAAAAAATAAGAACACAAAAAAGGTATCTAG 8 1 GCF_001544255.1 1050935 + no 1 AAAAAAATAAGAAGGTAGCACCAATAACTTT 8 1 GCF_900638025.1 137037 - no 1 AAAAAAATAAGCTGGGCCGTTTGGGGAACGA 8 1 GCF_000742135.1 989338 - no 1 AAAAAAATAAGGGGAAATTATGGCAGGTAAT 8 1 GCF_001457655.1 883695 - no 1 AAAAAAATAAGTGAAAATCTATTTTCTGAAA 8 1 GCF_000392875.1 2823442 - no 1 AAAAAAATAATATTGTCCATTCTCCTAGCAA 8 1 GCF_001544255.1 173045 - no 1 AAAAAAATAATCAAAGGCCGGGGATTATACG 8 1 GCF_003697165.2 733341 - no 1 AAAAAAATACCCTGCGTGATGATGCGAGGTG 8 1 GCF_002950215.1 1422485 - no 1 AAAAAAATACTTGCCTTCGGGCTTATCTCAG 8 1 GCF_003697165.2 2823100 + no 1 AAAAAAATACTTGTTTGATTCTGTATTACGT 8 1 GCF_000392875.1 493472 + no 1 AAAAAAATAGAAAATGAGTCAACACCACTAT 8 1 GCF_006742205.1 1365300 + no 1 AAAAAAATAGAATTATATCGTGAACGTTTTG 8 1 GCF_009759685.1 2234982 + no 1 AAAAAAATAGAGGATTAAATGCTAATTCATA 8 1 GCF_001457655.1 671915 + no 1 AAAAAAATAGTATAAATCCGCCATATAAAAT 8 1 GCF_001457655.1 1222761 - no -
Specify the mask.
$ lexicmap utils kmers --quiet -d demo.lmi/ --mask 12345 | head -n 20 | csvtk pretty -t mask kmer prefix number ref pos strand reversed ----- ------------------------------- ------ ------ --------------- ------- ------ -------- 12345 GCTGCACAAAGTACGATTACGATGCAAGCCC 8 1 GCF_002949675.1 716651 + no 12345 GCTGCACAACAAACGATTGTTGGTGAAATTT 8 1 GCF_000392875.1 836578 - no 12345 GCTGCACAACAACATGATAGTGTGAAATTAG 8 1 GCF_001027105.1 1150856 + no 12345 GCTGCACAACAGGCTGCGGCTGGTGTTGCGG 8 1 GCF_000742135.1 4128289 - no 12345 GCTGCACAACCAGGCAGAAAAAATAATGGGA 8 1 GCF_002950215.1 3009005 - no 12345 GCTGCACAACCTTTCCACAAGCCGTAAAACC 8 1 GCF_000006945.2 4306623 - no 12345 GCTGCACAACGATTAGAAAAAATGGGGTACG 8 1 GCF_001544255.1 2041481 - no 12345 GCTGCACAACTATCCCAATGCCGAGGTGGAA 8 1 GCF_000017205.1 5101754 + no 12345 GCTGCACAAGCACCCGGCCGTGGCCCTGGCG 8 1 GCF_000017205.1 1257468 + no 12345 GCTGCACAAGCGCTCGGTTTAGAGCAAACAC 8 1 GCF_009759685.1 1232954 - no 12345 GCTGCACAAGGGGCCACTTTCGTACATCGTC 8 1 GCF_000742135.1 3888020 + yes 12345 GCTGCACAAGTACCTGCTGGCCTACGCCTCG 8 1 GCF_000017205.1 1166094 + no 12345 GCTGCACAAGTTGCAAAACAGCTGATTAAGG 8 1 GCF_000392875.1 908172 + no 12345 GCTGCACAATATCGATTTGAACATTGCTCAG 8 1 GCF_003697165.2 3212441 + no 12345 GCTGCACAATATTTCATAATGACTTACGGCA 8 1 GCF_002950215.1 3443237 + no 12345 GCTGCACAATCCGCTGGGCTGGGTGCTCAAC 8 1 GCF_000742135.1 1083211 - no 12345 GCTGCACAATCGCCAGCCCCAGCCCTGTGCC 8 1 GCF_000006945.2 3658390 + no 12345 GCTGCACAATTACCACGTGAATTATTTGAAG 8 1 GCF_900638025.1 304434 - no 12345 GCTGCACAATTGCCAGCCCTAATCCCGTGCC 8 1 GCF_002950215.1 2671971 + no“reversed” means means if the k-mer is reversed for suffix matching. E.g.,
GCTGCACAAGGGGCCACTTTCGTACATCGTCis reversed, so you need to reverse it before searching in the genome.$ seqkit locate -p $(echo GCTGCACAAGGGGCCACTTTCGTACATCGTC | rev) refs/GCF_000742135.1.fa.gz -M | csvtk pretty -t seqID patternName pattern strand start end ------------- ------------------------------- ------------------------------- ------ ------- ------- NZ_KN046818.1 CTGCTACATGCTTTCACCGGGGAACACGTCG CTGCTACATGCTTTCACCGGGGAACACGTCG + 3888020 3888050 -
For all masks. The result might be very big, therefore, writing to gzip format is recommended.
$ lexicmap utils kmers -d demo.lmi/ --mask 0 -o kmers.tsv.gz $ zcat kmers.tsv.gz | csvtk freq -t -f mask -nr | head -n 10 mask frequency 8206 700 16230 636 4974 625 14979 620 11723 619 12043 593 12 589 18 589 17491 584a faster way
seq 1 $(lexicmap utils masks -d demo.lmi/ --quiet | wc -l) \ | rush --eta 'echo -e {}"\t"$(lexicmap utils kmers -d demo.lmi/ -m {} -f --quiet | csvtk nrow)' \ | csvtk add-header -t -n mask,seeds \ | csvtk sort -t -k seeds:nr \ | head -n 10 -
Lengths of shared prefixes between probes and captured k-mers.
zcat kmers.tsv.gz \ | csvtk grep -t -f reversed -p no \ | csvtk plot hist -t -f prefix -o prefix.hist.png \ --xlab "length of common prefixes between captured k-mers and masks"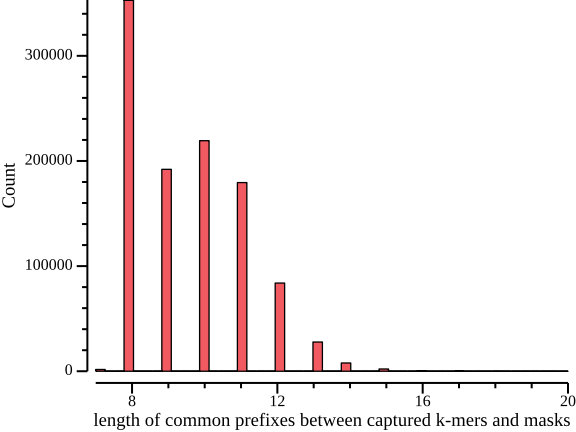
The output (TSV format) is formatted with csvtk pretty.