TaxonKit - A Practical and Efficient NCBI Taxonomy ToolkitLink
- Documents: https://bioinf.shenwei.me/taxonkit (Usage&Examples, Tutorial, 中文介绍)
- Source code: https://github.com/shenwei356/taxonkit
- Latest version:
- Please cite: https://doi.org/10.1016/j.jgg.2021.03.006
- pytaxonkit, Python bindings for TaxonKit.
Related projects:
- Taxid-Changelog: Tracking all changes of TaxIds, including deletion, new adding, merge, reuse, and rank/name changes.
- GTDB taxdump: GTDB taxonomy taxdump files with trackable TaxIds.
- ICTV taxdump: NCBI-style taxdump files for International Committee on Taxonomy of Viruses (ICTV)
Table of ContentsLink
- Features
- Subcommands
- Benchmark
- Dataset
- Installation
- Command-line completion
- Citation
- Contact
- License
FeaturesLink
- Easy to install (download)
- Statically linked executable binaries for multiple platforms (Linux/Windows/macOS, amd64/arm64)
- Light weight and out-of-the-box, no dependencies, no compilation, no configuration
- No database building, just download NCBI taxonomy data and uncompress to
$HOME/.taxonkit
- Easy to use (usages and examples)
- Supporting bash-completion
- Fast (see benchmark), multiple-CPUs supported, most operations cost 2-10s.
- Detailed usages and examples
- Supporting STDIN and (gzipped) input/output file, easily integrated in pipe
- Versatile commands
- Usage and examples
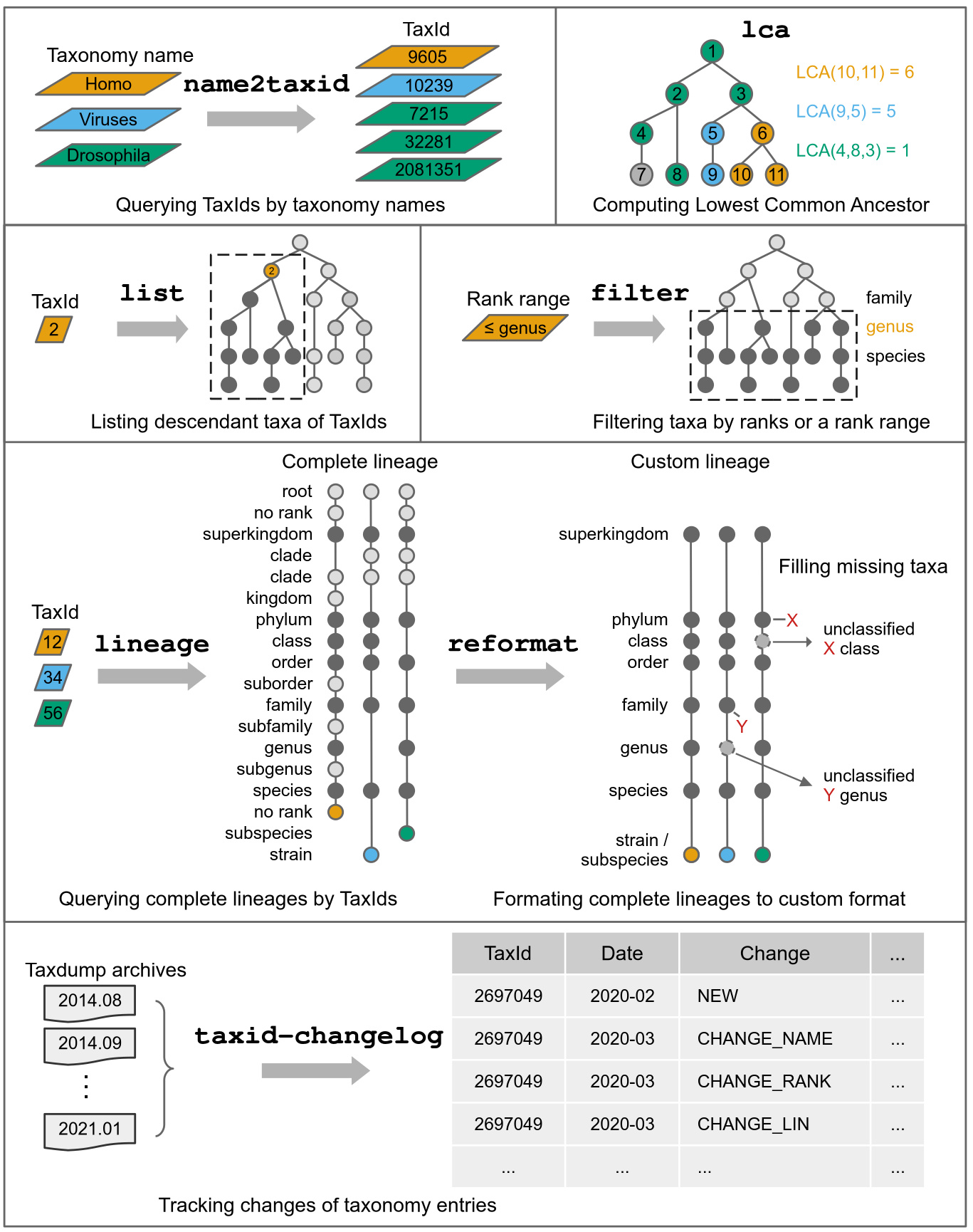
- Featured command: tracking monthly changelog of all TaxIds
- Featured command: reformating lineage into format of seven-level ("superkingdom/kingdom, phylum, class, order, family, genus, species", and even all possible ranks
- Featured command: filtering taxiDs by a rank range, e.g., at or below genus rank.
- Featured command: Create NCBI-style taxdump files for custom taxonomy, e.g., GTDB and ICTV
SubcommandsLink
| Subcommand | Function |
|---|---|
list |
List taxonomic subtrees (TaxIds) bellow given TaxIds |
lineage |
Query taxonomic lineage of given TaxIds |
reformat |
Reformat lineage in canonical ranks |
reformat2* |
Reformat lineage in chosen ranks, allowing more ranks than 'reformat' |
name2taxid |
Convert taxon names to TaxIds |
filter |
Filter TaxIds by taxonomic rank range |
lca |
Compute lowest common ancestor (LCA) for TaxIds |
taxid-changelog |
Create TaxId changelog from dump archives |
profile2cami* |
Convert metagenomic profile table to CAMI format |
cami-filter* |
Remove taxa of given TaxIds and their descendants in CAMI metagenomic profile |
create-taxdump* |
Create NCBI-style taxdump files for custom taxonomy, e.g., GTDB and ICTV |
Note: *New commands since the publication.
BenchmarkLink
- Getting complete lineage for given TaxIds (this plot is very old).
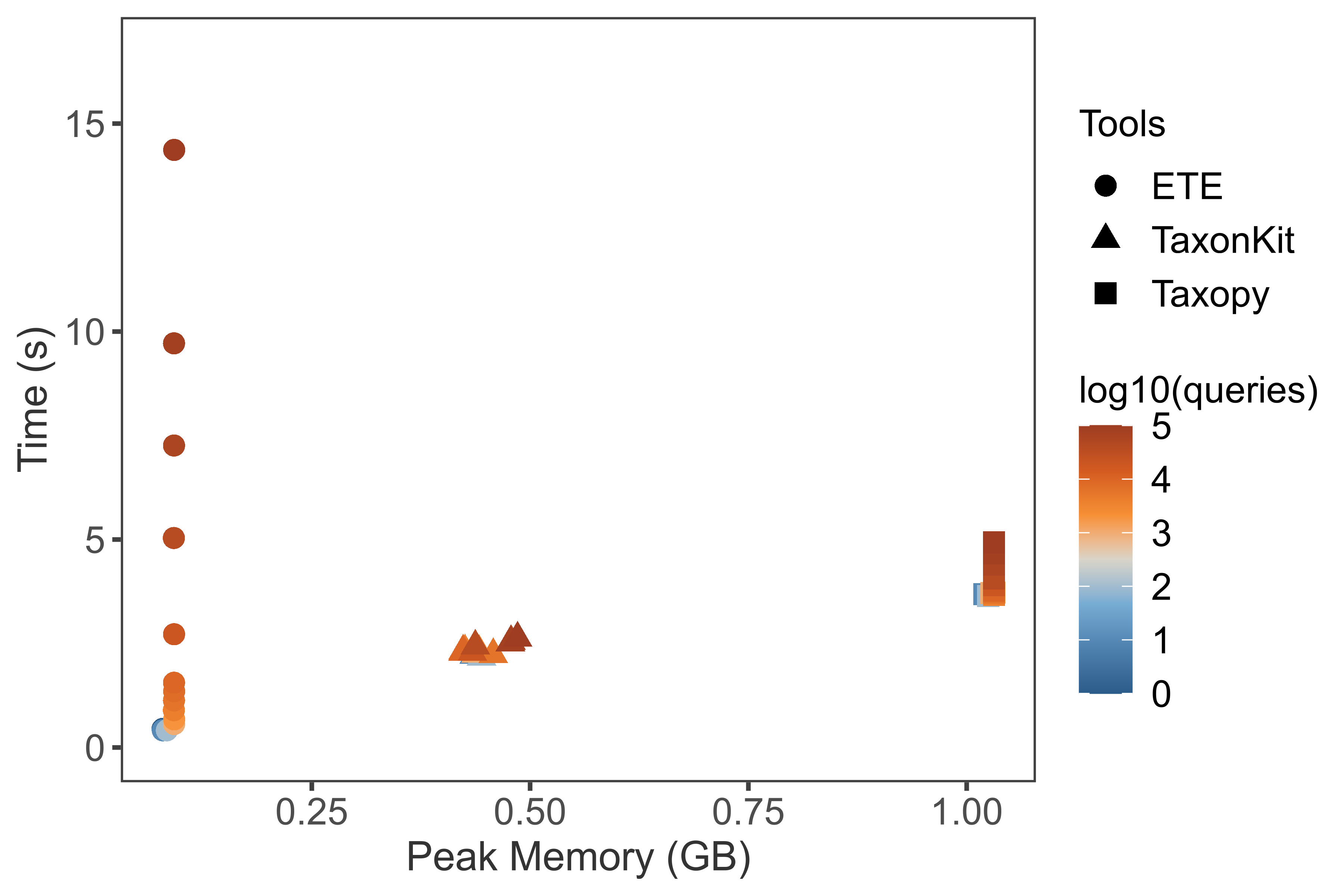
Versions: ETE=3.1.2, taxopy=0.5.0 (faster since 0.6.0), TaxonKit=0.7.2.
DatasetLink
- Download and uncompress
taxdump.tar.gz: https://ftp.ncbi.nih.gov/pub/taxonomy/taxdump.tar.gz - Copy
names.dmp,nodes.dmp,delnodes.dmpandmerged.dmpto data directory:$HOME/.taxonkit, e.g.,/home/shenwei/.taxonkit, - Optionally copy to some other directories, and later you can refer to using flag
--data-dir, or environment variableTAXONKIT_DB.
All-in-one command:
wget -c https://ftp.ncbi.nih.gov/pub/taxonomy/taxdump.tar.gz
tar -zxvf taxdump.tar.gz
mkdir -p $HOME/.taxonkit
cp names.dmp nodes.dmp delnodes.dmp merged.dmp $HOME/.taxonkit
Update dataset: Simply re-download the taxdump files, uncompress and override old ones.
InstallationLink
Go to Download Page for more download options and changelogs.
TaxonKit is implemented in Go programming language,
executable binary files for most popular operating systems are freely available
in release page.
Method 1: Download binaries (latest stable/dev version)Link
Just download compressed
executable file of your operating system,
and uncompress it with tar -zxvf *.tar.gz command or other tools.
And then:
-
For Linux-like systems
-
If you have root privilege simply copy it to
/usr/local/bin:sudo cp taxonkit /usr/local/bin/ -
Or copy to anywhere in the environment variable
PATH:mkdir -p $HOME/bin/; cp taxonkit $HOME/bin/
-
-
For Windows, just copy
taxonkit.exetoC:\WINDOWS\system32.
Method 2: Install via conda (latest stable version) 

 Link
Link
conda install -c bioconda taxonkit
Method 3: Install via homebrew (out of date)Link
brew install brewsci/bio/taxonkit
Method 4: Compile from source (latest stable/dev version)Link
-
wget https://go.dev/dl/go1.24.1.linux-amd64.tar.gz tar -zxf go1.24.1.linux-amd64.tar.gz -C $HOME/ # or # echo "export PATH=$PATH:$HOME/go/bin" >> ~/.bashrc # source ~/.bashrc export PATH=$PATH:$HOME/go/bin -
Compile TaxonKit
# ------------- the latest stable version ------------- go get -v -u github.com/shenwei356/taxonkit/taxonkit # The executable binary file is located in: # ~/go/bin/taxonkit # You can also move it to anywhere in the $PATH mkdir -p $HOME/bin cp ~/go/bin/taxonkit $HOME/bin/ # --------------- the development version -------------- git clone https://github.com/shenwei356/taxonkit cd taxonkit/taxonkit/ go build # The executable binary file is located in: # ./taxonkit # You can also move it to anywhere in the $PATH mkdir -p $HOME/bin cp ./taxonkit $HOME/bin/
Bash-completionLink
Supported shell: bash|zsh|fish|powershell
Bash:
# generate completion shell
taxonkit genautocomplete --shell bash
# configure if never did.
# install bash-completion if the "complete" command is not found.
echo "for bcfile in ~/.bash_completion.d/* ; do source \$bcfile; done" >> ~/.bash_completion
echo "source ~/.bash_completion" >> ~/.bashrc
Zsh:
# generate completion shell
taxonkit genautocomplete --shell zsh --file ~/.zfunc/_taxonkit
# configure if never did
echo 'fpath=( ~/.zfunc "${fpath[@]}" )' >> ~/.zshrc
echo "autoload -U compinit; compinit" >> ~/.zshrc
fish:
taxonkit genautocomplete --shell fish --file ~/.config/fish/completions/taxonkit.fish
CitationLink
If you use TaxonKit in your work, please cite:
Shen, W., Ren, H., TaxonKit: a practical and efficient NCBI Taxonomy toolkit, Journal of Genetics and Genomics, https://doi.org/10.1016/j.jgg.2021.03.006
ContactLink
Create an issue to report bugs, propose new functions or ask for help.





