Usage and ExamplesLink
Table of Contents
- Usage and Examples
- Before use
- taxonkit
- list
- lineage
- reformat
- reformat2
- name2taxid
- filter
- lca
- taxid-changelog
- profile2cami
- cami-filter
- create-taxdump
- genautocomplete
Before useLink
- Download and uncompress
taxdump.tar.gz: ftp://ftp.ncbi.nih.gov/pub/taxonomy/taxdump.tar.gz - Copy
names.dmp,nodes.dmp,delnodes.dmpandmerged.dmpto data directory:$HOME/.taxonkit, e.g.,/home/shenwei/.taxonkit, - Optionally copy to some other directories, and later you can refer to using flag
--data-dir, or environment variableTAXONKIT_DB.
All-in-one command:
wget -c ftp://ftp.ncbi.nih.gov/pub/taxonomy/taxdump.tar.gz
tar -zxvf taxdump.tar.gz
mkdir -p $HOME/.taxonkit
cp names.dmp nodes.dmp delnodes.dmp merged.dmp $HOME/.taxonkit
Update dataset: Simply re-download the taxdump files, uncompress and override old ones.
taxonkitLink
TaxonKit - A Practical and Efficient NCBI Taxonomy Toolkit
Version: 0.20.0
Author: Wei Shen <shenwei356@gmail.com>
Source code: https://github.com/shenwei356/taxonkit
Documents : https://bioinf.shenwei.me/taxonkit
Citation : https://www.sciencedirect.com/science/article/pii/S1673852721000837
Dataset:
Please download and uncompress "taxdump.tar.gz":
http://ftp.ncbi.nih.gov/pub/taxonomy/taxdump.tar.gz
and copy "names.dmp", "nodes.dmp", "delnodes.dmp" and "merged.dmp" to data directory:
"/home/shenwei/.taxonkit"
or some other directory, and later you can refer to using flag --data-dir,
or environment variable TAXONKIT_DB.
When environment variable TAXONKIT_DB is set, explicitly setting --data-dir will
overide the value of TAXONKIT_DB.
Usage:
taxonkit [command]
Available Commands:
cami-filter Remove taxa of given TaxIds and their descendants in CAMI metagenomic profile
create-taxdump Create NCBI-style taxdump files for custom taxonomy, e.g., GTDB and ICTV
filter Filter TaxIds by taxonomic rank range
genautocomplete generate shell autocompletion script (bash|zsh|fish|powershell)
lca Compute lowest common ancestor (LCA) for TaxIds
lineage Query taxonomic lineage of given TaxIds
list List taxonomic subtrees of given TaxIds
name2taxid Convert taxon names to TaxIds
profile2cami Convert metagenomic profile table to CAMI format
reformat Reformat lineage in canonical ranks
reformat2 Reformat lineage in chosen ranks, allowing more ranks than 'reformat'
taxid-changelog Create TaxId changelog from dump archives
version print version information and check for update
Flags:
--data-dir string directory containing nodes.dmp and names.dmp (default "/home/shenwei/.taxonkit")
-h, --help help for taxonkit
--line-buffered use line buffering on output, i.e., immediately writing to stdin/file for
every line of output
-o, --out-file string out file ("-" for stdout, suffix .gz for gzipped out) (default "-")
-j, --threads int number of CPUs. 4 is enough (default 4)
--verbose print verbose information
listLink
Usage
List taxonomic subtrees of given TaxIds
Attention:
1. When multiple taxids are given, the output may contain duplicated records
if some taxids are descendants of others.
Examples:
$ taxonkit list --ids 9606 -n -r --indent " "
9606 [species] Homo sapiens
63221 [subspecies] Homo sapiens neanderthalensis
741158 [subspecies] Homo sapiens subsp. 'Denisova'
$ taxonkit list --ids 9606 --indent ""
9606
63221
741158
# from stdin
echo 9606 | taxonkit list
# from file
taxonkit list <(echo 9606)
Usage:
taxonkit list [flags]
Flags:
-h, --help help for list
-i, --ids string TaxId(s), multiple values should be separated by comma
-I, --indent string indent (default " ")
-J, --json output in JSON format. you can save the result in file with suffix ".json" and
open with modern text editor
-n, --show-name output scientific name
-r, --show-rank output rank
Examples
-
Default usage.
$ taxonkit list --ids 9605,239934 9605 9606 63221 741158 1425170 2665952 2665953 239934 239935 349741 512293 512294 1131822 1262691 1263034 1679444 2608915 1131336 ... -
Removing indent. The list could be used to extract sequences from BLAST database with
blastdbcmd(see tutorial)$ taxonkit list --ids 9605,239934 --indent "" 9605 9606 63221 741158 1425170 2665952 2665953 239934 239935 349741 512293 512294 1131822 1262691 1263034 1679444 ...Performance: Time and memory usage for whole taxon tree:
$ # emptying the buffers cache $ su -c "free && sync && echo 3 > /proc/sys/vm/drop_caches && free" $ memusg -t taxonkit list --ids 1 --indent "" --verbose > t0.txt 21:05:01.782 [INFO] parsing merged file: /home/shenwei/.taxonkit/names.dmp 21:05:01.782 [INFO] parsing names file: /home/shenwei/.taxonkit/names.dmp 21:05:01.782 [INFO] parsing delnodes file: /home/shenwei/.taxonkit/names.dmp 21:05:01.816 [INFO] 61023 merged nodes parsed 21:05:01.889 [INFO] 437929 delnodes parsed 21:05:03.178 [INFO] 2303979 names parsed elapsed time: 3.290s peak rss: 742.77 MB -
Adding names
$ taxonkit list --show-rank --show-name --indent " " --ids 9605,239934 9605 [genus] Homo 9606 [species] Homo sapiens 63221 [subspecies] Homo sapiens neanderthalensis 741158 [subspecies] Homo sapiens subsp. 'Denisova' 1425170 [species] Homo heidelbergensis 2665952 [no rank] environmental samples 2665953 [species] Homo sapiens environmental sample 239934 [genus] Akkermansia 239935 [species] Akkermansia muciniphila 349741 [strain] Akkermansia muciniphila ATCC BAA-835 512293 [no rank] environmental samples 512294 [species] uncultured Akkermansia sp. 1131822 [species] uncultured Akkermansia sp. SMG25 1262691 [species] Akkermansia sp. CAG:344 1263034 [species] Akkermansia muciniphila CAG:154 1679444 [species] Akkermansia glycaniphila 2608915 [no rank] unclassified Akkermansia 1131336 [species] Akkermansia sp. KLE1605 1574264 [species] Akkermansia sp. KLE1797 ...Performance: Time and memory usage for whole taxonomy tree:
$ # emptying the buffers cache $ su -c "free && sync && echo 3 > /proc/sys/vm/drop_caches && free" $ memusg -t taxonkit list --show-rank --show-name --ids 1 > t1.txt elapsed time: 5.341s peak rss: 1.04 GB -
Output in JSON format, you can easily collapse and uncollapse taxonomy tree in modern text editor.
$ taxonkit list --show-rank --show-name --indent " " --ids 9605,239934 --json { "9605 [genus] Homo": { "9606 [species] Homo sapiens": { "63221 [subspecies] Homo sapiens neanderthalensis": { }, "741158 [subspecies] Homo sapiens subsp. 'Denisova'": { } }, "1425170 [species] Homo heidelbergensis": { } }, "239934 [genus] Akkermansia": { "239935 [species] Akkermansia muciniphila": { "349741 [no rank] Akkermansia muciniphila ATCC BAA-835": { } }, "512293 [no rank] environmental samples": { "512294 [species] uncultured Akkermansia sp.": { }, "1131822 [species] uncultured Akkermansia sp. SMG25": { }, "1262691 [species] Akkermansia sp. CAG:344": { }, "1263034 [species] Akkermansia muciniphila CAG:154": { } }, "1679444 [species] Akkermansia glycaniphila": { }, "2608915 [no rank] unclassified Akkermansia": { "1131336 [species] Akkermansia sp. KLE1605": { }, "1574264 [species] Akkermansia sp. KLE1797": { }, "1574265 [species] Akkermansia sp. KLE1798": { }, "1638783 [species] Akkermansia sp. UNK.MGS-1": { }, "1755639 [species] Akkermansia sp. MC_55": { } } } }Snapshot of taxonomy (taxid 1) in kate:
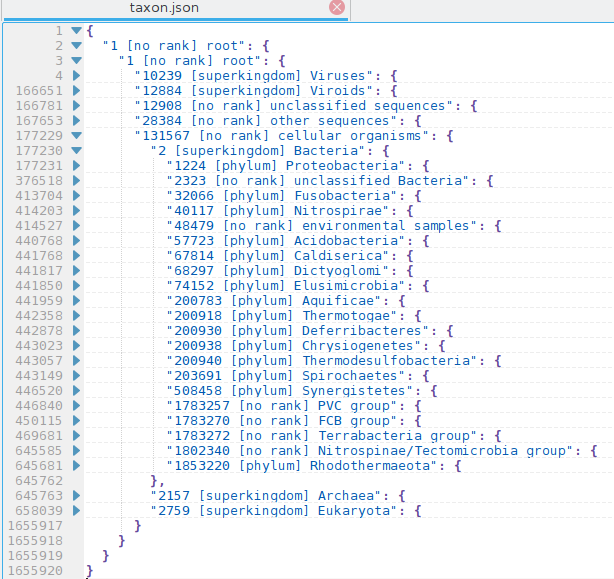
lineageLink
Usage
Query taxonomic lineage of given TaxIds
Input:
- List of TaxIds, one TaxId per line.
- Or tab-delimited format, please specify TaxId field
with flag -i/--taxid-field (default 1).
- Supporting (gzipped) file or STDIN.
Output:
1. Input line data.
2. (Optional) Status code (-c/--show-status-code), values:
- "-1" for queries not found in whole database.
- "0" for deleted TaxIds, provided by "delnodes.dmp".
- New TaxIds for merged TaxIds, provided by "merged.dmp".
- Taxids for these found in "nodes.dmp".
3. Lineage, delimiter can be changed with flag -d/--delimiter.
4. (Optional) TaxIds taxons in the lineage (-t/--show-lineage-taxids)
5. (Optional) Name (-n/--show-name)
6. (Optional) Rank (-r/--show-rank)
Filter out invalid and deleted taxids, and replace merged
taxids with new ones:
# input is one-column-taxid
$ taxonkit lineage -c taxids.txt \
| awk '$2>0' \
| cut -f 2-
# taxids are in 3rd field in a 4-columns tab-delimited file,
# for $5, where 5 = 4 + 1.
$ cat input.txt \
| taxonkit lineage -c -i 3 \
| csvtk filter2 -H -t -f '$5>0' \
| csvtk -H -t cut -f -3
Usage:
taxonkit lineage [flags]
Flags:
-d, --delimiter string field delimiter in lineage (default ";")
-h, --help help for lineage
-L, --no-lineage do not show lineage, when user just want names or/and ranks
-R, --show-lineage-ranks appending ranks of all levels
-t, --show-lineage-taxids appending lineage consisting of taxids
-n, --show-name appending scientific name
-r, --show-rank appending rank of taxids
-c, --show-status-code show status code before lineage
-i, --taxid-field int field index of taxid. input data should be tab-separated (default 1)
Examples
-
Full lineage:
# note that 123124124 is a fake taxid, 3 was deleted, 92489,1458427 were merged $ cat taxids.txt 9606 9913 376619 349741 239935 314101 11932 1327037 123124124 3 92489 1458427 $ taxonkit lineage taxids.txt | tee lineage.txt 19:22:13.077 [WARN] taxid 92489 was merged into 796334 19:22:13.077 [WARN] taxid 1458427 was merged into 1458425 19:22:13.077 [WARN] taxid 123124124 not found 19:22:13.077 [WARN] taxid 3 was deleted 9606 cellular organisms;Eukaryota;Opisthokonta;Metazoa;Eumetazoa;Bilateria;Deuterostomia;Chordata;Craniata;Vertebrata;Gnathostomata;Teleostomi;Euteleostomi;Sarcopterygii;Dipnotetrapodomorpha;Tetrapoda;Amniota;Mammalia;Theria;Eutheria;Boreoeutheria;Euarchontoglires;Primates;Haplorrhini;Simiiformes;Catarrhini;Hominoidea;Hominidae;Homininae;Homo;Homo sapiens 9913 cellular organisms;Eukaryota;Opisthokonta;Metazoa;Eumetazoa;Bilateria;Deuterostomia;Chordata;Craniata;Vertebrata;Gnathostomata;Teleostomi;Euteleostomi;Sarcopterygii;Dipnotetrapodomorpha;Tetrapoda;Amniota;Mammalia;Theria;Eutheria;Boreoeutheria;Laurasiatheria;Artiodactyla;Ruminantia;Pecora;Bovidae;Bovinae;Bos;Bos taurus 376619 cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria;Thiotrichales;Francisellaceae;Francisella;Francisella tularensis;Francisella tularensis subsp. holarctica;Francisella tularensis subsp. holarctica LVS 349741 cellular organisms;Bacteria;PVC group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila;Akkermansia muciniphila ATCC BAA-835 239935 cellular organisms;Bacteria;PVC group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila 314101 cellular organisms;Bacteria;environmental samples;uncultured murine large bowel bacterium BAC 54B 11932 Viruses;Riboviria;Pararnavirae;Artverviricota;Revtraviricetes;Ortervirales;Retroviridae;unclassified Retroviridae;Intracisternal A-particles;Mouse Intracisternal A-particle 1327037 Viruses;Duplodnaviria;Heunggongvirae;Uroviricota;Caudoviricetes;Caudovirales;Siphoviridae;unclassified Siphoviridae;Croceibacter phage P2559Y 123124124 3 92489 cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacterales;Erwiniaceae;Erwinia;Erwinia oleae 1458427 cellular organisms;Bacteria;Proteobacteria;Betaproteobacteria;Burkholderiales;Comamonadaceae;Serpentinomonas;Serpentinomonas raicheisms;Bacteria;Proteobacteria;Betaproteobacteria;Burkholderiales;Comamonadaceae;Serpentinomonas;Serpentinomonas raichei # wrapped table with csvtk pretty (>v0.26.0) $ taxonkit lineage taxids.txt | csvtk pretty -Ht -x ';' -W 70 -S bold ┏━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ 9606 ┃ cellular organisms;Eukaryota;Opisthokonta;Metazoa;Eumetazoa;Bilateria; ┃ ┃ ┃ Deuterostomia;Chordata;Craniata;Vertebrata;Gnathostomata;Teleostomi; ┃ ┃ ┃ Euteleostomi;Sarcopterygii;Dipnotetrapodomorpha;Tetrapoda;Amniota; ┃ ┃ ┃ Mammalia;Theria;Eutheria;Boreoeutheria;Euarchontoglires;Primates; ┃ ┃ ┃ Haplorrhini;Simiiformes;Catarrhini;Hominoidea;Hominidae;Homininae; ┃ ┃ ┃ Homo;Homo sapiens ┃ ┣━━━━━━━━━╋━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┫ ┃ 9913 ┃ cellular organisms;Eukaryota;Opisthokonta;Metazoa;Eumetazoa;Bilateria; ┃ ┃ ┃ Deuterostomia;Chordata;Craniata;Vertebrata;Gnathostomata;Teleostomi; ┃ ┃ ┃ Euteleostomi;Sarcopterygii;Dipnotetrapodomorpha;Tetrapoda;Amniota; ┃ ┃ ┃ Mammalia;Theria;Eutheria;Boreoeutheria;Laurasiatheria;Artiodactyla; ┃ ┃ ┃ Ruminantia;Pecora;Bovidae;Bovinae;Bos;Bos taurus ┃ ┣━━━━━━━━━╋━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┫ ┃ 376619 ┃ cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria; ┃ ┃ ┃ Thiotrichales;Francisellaceae;Francisella;Francisella tularensis; ┃ ┃ ┃ Francisella tularensis subsp. holarctica; ┃ ┃ ┃ Francisella tularensis subsp. holarctica LVS ┃ ┣━━━━━━━━━╋━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┫ ┃ 349741 ┃ cellular organisms;Bacteria;PVC group;Verrucomicrobia; ┃ ┃ ┃ Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia; ┃ ┃ ┃ Akkermansia muciniphila;Akkermansia muciniphila ATCC BAA-835 ┃ ┣━━━━━━━━━╋━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┫ ┃ 239935 ┃ cellular organisms;Bacteria;PVC group;Verrucomicrobia; ┃ ┃ ┃ Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia; ┃ ┃ ┃ Akkermansia muciniphila ┃ ┣━━━━━━━━━╋━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┫ ┃ 314101 ┃ cellular organisms;Bacteria;environmental samples; ┃ ┃ ┃ uncultured murine large bowel bacterium BAC 54B ┃ ┣━━━━━━━━━╋━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┫ ┃ 11932 ┃ Viruses;Riboviria;Pararnavirae;Artverviricota;Revtraviricetes; ┃ ┃ ┃ Ortervirales;Retroviridae;unclassified Retroviridae; ┃ ┃ ┃ Intracisternal A-particles;Mouse Intracisternal A-particle ┃ ┣━━━━━━━━━╋━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┫ ┃ 1327037 ┃ Viruses;Duplodnaviria;Heunggongvirae;Uroviricota;Caudoviricetes; ┃ ┃ ┃ Caudovirales;Siphoviridae;unclassified Siphoviridae; ┃ ┃ ┃ Croceibacter phage P2559Y ┃ ┣━━━━━━━━━╋━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┫ ┃ 92489 ┃ cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria; ┃ ┃ ┃ Enterobacterales;Erwiniaceae;Erwinia;Erwinia oleae ┃ ┣━━━━━━━━━╋━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┫ ┃ 1458427 ┃ cellular organisms;Bacteria;Proteobacteria;Betaproteobacteria; ┃ ┃ ┃ Burkholderiales;Comamonadaceae;Serpentinomonas; ┃ ┃ ┃ Serpentinomonas raichei ┃ ┗━━━━━━━━━┻━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┛ -
Speed.
$ time echo 9606 | taxonkit lineage 9606 cellular organisms;Eukaryota;Opisthokonta;Metazoa;Eumetazoa;Bilateria;Deuterostomia;Chordata;Craniata;Vertebrata;Gnathostomata;Teleostomi;Euteleostomi;Sarcopterygii;Dipnotetrapodomorpha;Tetrapoda;Amniota;Mammalia;Theria;Eutheria;Boreoeutheria;Euarchontoglires;Primates;Haplorrhini;Simiiformes;Catarrhini;Hominoidea;Hominidae;Homininae;Homo;Homo sapiens real 0m1.190s user 0m2.365s sys 0m0.170s # all TaxIds $ time taxonkit list --ids 1 --indent "" | taxonkit lineage > t real 0m4.249s user 0m16.418s sys 0m1.221s -
Checking deleted or merged taxids
$ taxonkit lineage --show-status-code taxids.txt | tee lineage.withcode.txt # valid $ cat lineage.withcode.txt | awk '$2 > 0' | cut -f 1,2 9606 9606 9913 9913 376619 376619 349741 349741 239935 239935 314101 314101 11932 11932 1327037 1327037 92489 796334 1458427 1458425 # merged $ cat lineage.withcode.txt | awk '$2 > 0 && $2 != $1' | cut -f 1,2 92489 796334 1458427 1458425 # deleted $ cat lineage.withcode.txt | awk '$2 == 0' | cut -f 1 3 # invalid $ cat lineage.withcode.txt | awk '$2 < 0' | cut -f 1 123124124 -
Filter out invalid and deleted taxids, and replace merged taxids with new ones, you may install csvtk.
# input is one-column-taxid $ taxonkit lineage -c taxids.txt \ | awk '$2>0' \ | cut -f 2- # taxids are in 3rd field in a 4-columns tab-delimited file, # for $5, where 5 = 4 + 1. $ cat input.txt \ | taxonkit lineage -c -i 3 \ | csvtk filter2 -H -t -f '$5>0' \ | csvtk -H -t cut -f -3 -
Only show name and rank.
$ taxonkit lineage -r -n -L taxids.txt \ | csvtk pretty -H -t 9606 Homo sapiens species 9913 Bos taurus species 376619 Francisella tularensis subsp. holarctica LVS strain 349741 Akkermansia muciniphila ATCC BAA-835 strain 239935 Akkermansia muciniphila species 314101 uncultured murine large bowel bacterium BAC 54B species 11932 Mouse Intracisternal A-particle species 1327037 Croceibacter phage P2559Y species 123124124 3 92489 Erwinia oleae species 1458427 Serpentinomonas raichei species -
Show lineage consisting of taxids:
$ taxonkit lineage -t taxids.txt 9606 cellular organisms;Eukaryota;Opisthokonta;Metazoa;Eumetazoa;Bilateria;Deuterostomia;Chordata;Craniata;Vertebrata;Gnathostomata;Teleostomi;Euteleostomi;Sarcopterygii;Dipnotetrapodomorpha;Tetrapoda;Amniota;Mammalia;Theria;Eutheria;Boreoeutheria;Euarchontoglires;Primates;Haplorrhini;Simiiformes;Catarrhini;Hominoidea;Hominidae;Homininae;Homo;Homo sapiens 131567;2759;33154;33208;6072;33213;33511;7711;89593;7742;7776;117570;117571;8287;1338369;32523;32524;40674;32525;9347;1437010;314146;9443;376913;314293;9526;314295;9604;207598;9605;9606 9913 cellular organisms;Eukaryota;Opisthokonta;Metazoa;Eumetazoa;Bilateria;Deuterostomia;Chordata;Craniata;Vertebrata;Gnathostomata;Teleostomi;Euteleostomi;Sarcopterygii;Dipnotetrapodomorpha;Tetrapoda;Amniota;Mammalia;Theria;Eutheria;Boreoeutheria;Laurasiatheria;Artiodactyla;Ruminantia;Pecora;Bovidae;Bovinae;Bos;Bos taurus 131567;2759;33154;33208;6072;33213;33511;7711;89593;7742;7776;117570;117571;8287;1338369;32523;32524;40674;32525;9347;1437010;314145;91561;9845;35500;9895;27592;9903;9913 376619 cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria;Thiotrichales;Francisellaceae;Francisella;Francisella tularensis;Francisella tularensis subsp. holarctica;Francisella tularensis subsp. holarctica LVS 131567;2;1224;1236;72273;34064;262;263;119857;376619 349741 cellular organisms;Bacteria;PVC group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila;Akkermansia muciniphila ATCC BAA-835 131567;2;1783257;74201;203494;48461;1647988;239934;239935;349741 239935 cellular organisms;Bacteria;PVC group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila 131567;2;1783257;74201;203494;48461;1647988;239934;239935 314101 cellular organisms;Bacteria;environmental samples;uncultured murine large bowel bacterium BAC 54B 131567;2;48479;314101 11932 Viruses;Riboviria;Pararnavirae;Artverviricota;Revtraviricetes;Ortervirales;Retroviridae;unclassified Retroviridae;Intracisternal A-particles;Mouse Intracisternal A-particle 10239;2559587;2732397;2732409;2732514;2169561;11632;35276;11749;11932 1327037 Viruses;Duplodnaviria;Heunggongvirae;Uroviricota;Caudoviricetes;Caudovirales;Siphoviridae;unclassified Siphoviridae;Croceibacter phage P2559Y 10239;2731341;2731360;2731618;2731619;28883;10699;196894;1327037 123124124 3 92489 cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacterales;Erwiniaceae;Erwinia;Erwinia oleae 131567;2;1224;1236;91347;1903409;551;796334 1458427 cellular organisms;Bacteria;Proteobacteria;Betaproteobacteria;Burkholderiales;Comamonadaceae;Serpentinomonas;Serpentinomonas raichei 131567;2;1224;28216;80840;80864;2490452;1458425or read taxids from STDIN:
$ cat taxids.txt | taxonkit lineage -
And ranks of all nodes:
$ echo 2697049 \ | taxonkit lineage -t -R \ | csvtk transpose -Ht 2697049 Viruses;Riboviria;Orthornavirae;Pisuviricota;Pisoniviricetes;Nidovirales;Cornidovirineae;Coronaviridae;Orthocoronavirinae;Betacoronavirus;Sarbecovirus;Severe acute respiratory syndrome-related coronavirus;Severe acute respiratory syndrome coronavirus 2 10239;2559587;2732396;2732408;2732506;76804;2499399;11118;2501931;694002;2509511;694009;2697049 superkingdom;clade;kingdom;phylum;class;order;suborder;family;subfamily;genus;subgenus;species;no rankAnother way to show lineage detail of a TaxId
$ echo 2697049 \ | taxonkit lineage -t \ | csvtk cut -Ht -f 3 \ | csvtk unfold -Ht -f 1 -s ";" \ | taxonkit lineage -r -n -L \ | csvtk cut -Ht -f 1,3,2 \ | csvtk pretty -H -t 10239 superkingdom Viruses 2559587 clade Riboviria 2732396 kingdom Orthornavirae 2732408 phylum Pisuviricota 2732506 class Pisoniviricetes 76804 order Nidovirales 2499399 suborder Cornidovirineae 11118 family Coronaviridae 2501931 subfamily Orthocoronavirinae 694002 genus Betacoronavirus 2509511 subgenus Sarbecovirus 694009 species Severe acute respiratory syndrome-related coronavirus 2697049 no rank Severe acute respiratory syndrome coronavirus 2
reformatLink
Usage
Reformat lineage in canonical ranks
Warning:
- 'taxonkit reformat2' is recommended since Match 2025 when NCBI made
big changes to ranks.
See more: https://ncbiinsights.ncbi.nlm.nih.gov/2025/02/27/new-ranks-ncbi-taxonomy/
Input:
- List of TaxIds or lineages, one record per line.
The lineage can be a complete lineage or only one taxonomy name.
- Or tab-delimited format.
Plese specify the lineage field with flag -i/--lineage-field (default 2).
Or specify the TaxId field with flag -I/--taxid-field (default 0),
which overrides -i/--lineage-field.
- Supporting (gzipped) file or STDIN.
Output:
1. Input line data.
2. Reformated lineage.
3. (Optional) TaxIds taxons in the lineage (-t/--show-lineage-taxids)
Ambiguous names:
- Some TaxIds have the same complete lineage, empty result is returned
by default. You can use the flag -a/--output-ambiguous-result to
return one possible result
Output format can be formated by flag --format, available placeholders:
{C}: cellular root
{a}: acellular root
{r}: realm
{d}: domain
{k}: superkingdom
{K}: kingdom
{p}: phylum
{c}: class
{o}: order
{f}: family
{g}: genus
{s}: species
{t}: subspecies/strain
{S}: subspecies
{T}: strain
When these're no nodes of rank "subspecies" nor "strain",
you can switch on -S/--pseudo-strain to use the node with lowest rank
as subspecies/strain name, if which rank is lower than "species".
This flag affects {t}, {S}, {T}.
Output format can contains some escape charactors like "\t".
Usage:
taxonkit reformat [flags]
Flags:
-P, --add-prefix add prefixes for all ranks, single prefix for a rank is defined
by flag --prefix-X
-d, --delimiter string field delimiter in input lineage (default ";")
-F, --fill-miss-rank fill missing rank with lineage information of the next higher rank
-f, --format string output format, placeholders of rank are needed (default
"{k};{p};{c};{o};{f};{g};{s}")
-h, --help help for reformat
-i, --lineage-field int field index of lineage. data should be tab-separated (default 2)
-r, --miss-rank-repl string replacement string for missing rank
-p, --miss-rank-repl-prefix string prefix for estimated taxon names (default "unclassified ")
-s, --miss-rank-repl-suffix string suffix for estimated taxon names. "rank" for rank name, "" for no
suffix (default "rank")
-R, --miss-taxid-repl string replacement string for missing taxid
-a, --output-ambiguous-result output one of the ambigous result
--prefix-C string prefix for cellular root, used along with flag -P/--add-prefix
(default "d__")
--prefix-K string prefix for kingdom, used along with flag -P/--add-prefix (default
"K__")
--prefix-S string prefix for subspecies, used along with flag -P/--add-prefix
(default "S__")
--prefix-T string prefix for strain, used along with flag -P/--add-prefix (default
"T__")
--prefix-a string prefix for acellular root, used along with flag -P/--add-prefix
(default "d__")
--prefix-c string prefix for class, used along with flag -P/--add-prefix (default "c__")
--prefix-d string prefix for domain, used along with flag -P/--add-prefix (default
"d__")
--prefix-f string prefix for family, used along with flag -P/--add-prefix (default
"f__")
--prefix-g string prefix for genus, used along with flag -P/--add-prefix (default "g__")
--prefix-k string prefix for superkingdom, used along with flag -P/--add-prefix
(default "k__")
--prefix-o string prefix for order, used along with flag -P/--add-prefix (default "o__")
--prefix-p string prefix for phylum, used along with flag -P/--add-prefix (default
"p__")
--prefix-r string prefix for realm, used along with flag -P/--add-prefix (default "r__")
--prefix-s string prefix for species, used along with flag -P/--add-prefix (default
"s__")
--prefix-t string prefix for subspecies/strain, used along with flag
-P/--add-prefix (default "t__")
-S, --pseudo-strain use the node with lowest rank as strain name, only if which rank
is lower than "species" and not "subpecies" nor "strain". It
affects {t}, {S}, {T}. This flag needs flag -F
-t, --show-lineage-taxids show corresponding taxids of reformated lineage
-I, --taxid-field int field index of taxid. input data should be tab-separated. it
overrides -i/--lineage-field
-T, --trim do not fill or add prefix for missing rank lower than current rank
Examples:
-
For version > 0.8.0,
reformataccept input of TaxIds via flag-I/--taxid-field.$ echo 239935 | taxonkit reformat -I 1 239935 Bacteria;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila $ echo 349741 | taxonkit reformat -I 1 -f "{k}|{p}|{c}|{o}|{f}|{g}|{s}|{t}" -F -t 349741 Bacteria|Verrucomicrobia|Verrucomicrobiae|Verrucomicrobiales|Akkermansiaceae|Akkermansia|Akkermansia muciniphila|Akkermansia muciniphila ATCC BAA-835 2|74201|203494|48461|1647988|239934|239935|349741 -
Example lineage (produced by:
taxonkit lineage taxids.txt | awk '$2!=""' > lineage.txt).$ cat lineage.txt 9606 cellular organisms;Eukaryota;Opisthokonta;Metazoa;Eumetazoa;Bilateria;Deuterostomia;Chordata;Craniata;Vertebrata;Gnathostomata;Teleostomi;Euteleostomi;Sarcopterygii;Dipnotetrapodomorpha;Tetrapoda;Amniota;Mammalia;Theria;Eutheria;Boreoeutheria;Euarchontoglires;Primates;Haplorrhini;Simiiformes;Catarrhini;Hominoidea;Hominidae;Homininae;Homo;Homo sapiens 9913 cellular organisms;Eukaryota;Opisthokonta;Metazoa;Eumetazoa;Bilateria;Deuterostomia;Chordata;Craniata;Vertebrata;Gnathostomata;Teleostomi;Euteleostomi;Sarcopterygii;Dipnotetrapodomorpha;Tetrapoda;Amniota;Mammalia;Theria;Eutheria;Boreoeutheria;Laurasiatheria;Artiodactyla;Ruminantia;Pecora;Bovidae;Bovinae;Bos;Bos taurus 376619 cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria;Thiotrichales;Francisellaceae;Francisella;Francisella tularensis;Francisella tularensis subsp. holarctica;Francisella tularensis subsp. holarctica LVS 349741 cellular organisms;Bacteria;PVC group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila;Akkermansia muciniphila ATCC BAA-835 239935 cellular organisms;Bacteria;PVC group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila 314101 cellular organisms;Bacteria;environmental samples;uncultured murine large bowel bacterium BAC 54B 11932 Viruses;Riboviria;Pararnavirae;Artverviricota;Revtraviricetes;Ortervirales;Retroviridae;unclassified Retroviridae;Intracisternal A-particles;Mouse Intracisternal A-particle 1327037 Viruses;Duplodnaviria;Heunggongvirae;Uroviricota;Caudoviricetes;Caudovirales;Siphoviridae;unclassified Siphoviridae;Croceibacter phage P2559Y 92489 cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacterales;Erwiniaceae;Erwinia;Erwinia oleae 1458427 cellular organisms;Bacteria;Proteobacteria;Betaproteobacteria;Burkholderiales;Comamonadaceae;Serpentinomonas;Serpentinomonas raichei -
Default output format (
"{k};{p};{c};{o};{f};{g};{s}").# reformated lineages are appended to the input data $ taxonkit reformat lineage.txt ... 239935 cellular organisms;Bacteria;PVC group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila Bacteria;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila ... $ $ taxonkit reformat lineage.txt | tee lineage.txt.reformat $ cut -f 1,3 lineage.txt.reformat 9606 Eukaryota;Chordata;Mammalia;Primates;Hominidae;Homo;Homo sapiens 9913 Eukaryota;Chordata;Mammalia;Artiodactyla;Bovidae;Bos;Bos taurus 376619 Bacteria;Proteobacteria;Gammaproteobacteria;Thiotrichales;Francisellaceae;Francisella;Francisella tularensis 349741 Bacteria;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila 239935 Bacteria;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila 314101 Bacteria;;;;;;uncultured murine large bowel bacterium BAC 54B 11932 Viruses;Artverviricota;Revtraviricetes;Ortervirales;Retroviridae;Intracisternal A-particles;Mouse Intracisternal A-particle 1327037 Viruses;Uroviricota;Caudoviricetes;Caudovirales;Siphoviridae;;Croceibacter phage P2559Y 92489 Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacterales;Erwiniaceae;Erwinia;Erwinia oleae 1458427 Bacteria;Proteobacteria;Betaproteobacteria;Burkholderiales;Comamonadaceae;Serpentinomonas;Serpentinomonas raichei # aligned $ cat lineage.txt \ | taxonkit reformat \ | csvtk -H -t cut -f 1,3 \ | csvtk -H -t sep -f 2 -s ';' -R \ | csvtk add-header -t -n taxid,kindom,phylum,class,order,family,genus,species \ | csvtk pretty -t taxid kindom phylum class order family genus species ------- --------- --------------- ------------------- ------------------ --------------- -------------------------- ----------------------------------------------- 9606 Eukaryota Chordata Mammalia Primates Hominidae Homo Homo sapiens 9913 Eukaryota Chordata Mammalia Artiodactyla Bovidae Bos Bos taurus 376619 Bacteria Proteobacteria Gammaproteobacteria Thiotrichales Francisellaceae Francisella Francisella tularensis 349741 Bacteria Verrucomicrobia Verrucomicrobiae Verrucomicrobiales Akkermansiaceae Akkermansia Akkermansia muciniphila 239935 Bacteria Verrucomicrobia Verrucomicrobiae Verrucomicrobiales Akkermansiaceae Akkermansia Akkermansia muciniphila 314101 Bacteria uncultured murine large bowel bacterium BAC 54B 11932 Viruses Artverviricota Revtraviricetes Ortervirales Retroviridae Intracisternal A-particles Mouse Intracisternal A-particle 1327037 Viruses Uroviricota Caudoviricetes Caudovirales Siphoviridae Croceibacter phage P2559Y 92489 Bacteria Proteobacteria Gammaproteobacteria Enterobacterales Erwiniaceae Erwinia Erwinia oleae 1458427 Bacteria Proteobacteria Betaproteobacteria Burkholderiales Comamonadaceae Serpentinomonas Serpentinomonas raichei -
And
subspecies/strain({t}),subspecies({S}), andstrain({T}) are also available.# default operation $ echo -ne "239935\n83333\n1408252\n2697049\n2605619\n" \ | taxonkit lineage -n -r \ | taxonkit reformat -f '{t};{S};{T}' \ | csvtk -H -t cut -f 1,4,3,5 \ | csvtk -H -t sep -f 4 -s ';' -R \ | csvtk -H -t add-header -n "taxid,rank,name,subspecies/strain,subspecies,strain" \ | csvtk pretty -t taxid rank name subspecies/strain subspecies strain ------- ---------- ----------------------------------------------- --------------------- --------------------- --------------------- 239935 species Akkermansia muciniphila 83333 strain Escherichia coli K-12 Escherichia coli K-12 Escherichia coli K-12 1408252 subspecies Escherichia coli R178 Escherichia coli R178 Escherichia coli R178 2697049 no rank Severe acute respiratory syndrome coronavirus 2 2605619 no rank Escherichia coli O16:H48 # fill missing ranks # see example below for -F/--fill-miss-rank # $ echo -ne "239935\n83333\n1408252\n2697049\n2605619\n" \ | taxonkit lineage -n -r \ | taxonkit reformat -f '{t};{S};{T}' --fill-miss-rank \ | csvtk -H -t cut -f 1,4,3,5 \ | csvtk -H -t sep -f 4 -s ';' -R \ | csvtk -H -t add-header -n "taxid,rank,name,subspecies/strain,subspecies,strain" \ | csvtk pretty -t taxid rank name subspecies/strain subspecies strain ------- ---------- ----------------------------------------------- ------------------------------------------------------------------------------------ ----------------------------------------------------------------------------- ------------------------------------------------------------------------- 239935 species Akkermansia muciniphila unclassified Akkermansia muciniphila subspecies/strain unclassified Akkermansia muciniphila subspecies unclassified Akkermansia muciniphila strain 83333 strain Escherichia coli K-12 Escherichia coli K-12 unclassified Escherichia coli subspecies Escherichia coli K-12 1408252 subspecies Escherichia coli R178 Escherichia coli R178 Escherichia coli R178 unclassified Escherichia coli R178 strain 2697049 no rank Severe acute respiratory syndrome coronavirus 2 unclassified Severe acute respiratory syndrome-related coronavirus subspecies/strain unclassified Severe acute respiratory syndrome-related coronavirus subspecies unclassified Severe acute respiratory syndrome-related coronavirus strain 2605619 no rank Escherichia coli O16:H48 unclassified Escherichia coli subspecies/strain unclassified Escherichia coli subspecies unclassified Escherichia coli strain -
When these's no nodes of rank "subspecies" nor "strain", you can switch
-S/--pseudo-strainto use the node with lowest rank as subspecies/strain name, if which rank is lower than "species". Recommend using v0.14.1 or later versions.$ echo -ne "239935\n83333\n1408252\n2697049\n2605619\n" \ | taxonkit lineage -n -r \ | taxonkit reformat -f '{t};{S};{T}' --pseudo-strain \ | csvtk -H -t cut -f 1,4,3,5 \ | csvtk -H -t sep -f 4 -s ';' -R \ | csvtk -H -t add-header -n "taxid,rank,name,subspecies/strain,subspecies,strain" \ | csvtk pretty -t taxid rank name subspecies/strain subspecies strain ------- ---------- ----------------------------------------------- ----------------------------------------------- ----------------------------------------------- ----------------------------------------------- 239935 species Akkermansia muciniphila 83333 strain Escherichia coli K-12 Escherichia coli K-12 Escherichia coli K-12 1408252 subspecies Escherichia coli R178 Escherichia coli R178 Escherichia coli R178 2697049 no rank Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 Severe acute respiratory syndrome coronavirus 2 2605619 no rank Escherichia coli O16:H48 Escherichia coli O16:H48 Escherichia coli O16:H48 Escherichia coli O16:H48 -
Add prefix (
-P/--add-prefix).$ cat lineage.txt \ | taxonkit reformat -P \ | csvtk -H -t cut -f 1,3 9606 k__Eukaryota;p__Chordata;c__Mammalia;o__Primates;f__Hominidae;g__Homo;s__Homo sapiens 9913 k__Eukaryota;p__Chordata;c__Mammalia;o__Artiodactyla;f__Bovidae;g__Bos;s__Bos taurus 376619 k__Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Thiotrichales;f__Francisellaceae;g__Francisella;s__Francisella tularensis 349741 k__Bacteria;p__Verrucomicrobia;c__Verrucomicrobiae;o__Verrucomicrobiales;f__Akkermansiaceae;g__Akkermansia;s__Akkermansia muciniphila 239935 k__Bacteria;p__Verrucomicrobia;c__Verrucomicrobiae;o__Verrucomicrobiales;f__Akkermansiaceae;g__Akkermansia;s__Akkermansia muciniphila 314101 k__Bacteria;p__;c__;o__;f__;g__;s__uncultured murine large bowel bacterium BAC 54B 11932 k__Viruses;p__Artverviricota;c__Revtraviricetes;o__Ortervirales;f__Retroviridae;g__Intracisternal A-particles;s__Mouse Intracisternal A-particle 1327037 k__Viruses;p__Uroviricota;c__Caudoviricetes;o__Caudovirales;f__Siphoviridae;g__;s__Croceibacter phage P2559Y 92489 k__Bacteria;p__Proteobacteria;c__Gammaproteobacteria;o__Enterobacterales;f__Erwiniaceae;g__Erwinia;s__Erwinia oleae 1458427 k__Bacteria;p__Proteobacteria;c__Betaproteobacteria;o__Burkholderiales;f__Comamonadaceae;g__Serpentinomonas;s__Serpentinomonas raichei -
Show corresponding taxids of reformated lineage (flag
-t/--show-lineage-taxids)$ cat lineage.txt \ | taxonkit reformat -t \ | csvtk -H -t cut -f 1,4 \ | csvtk -H -t sep -f 2 -s ';' -R \ | csvtk add-header -t -n taxid,kindom,phylum,class,order,family,genus,species \ | csvtk pretty -t taxid kindom phylum class order family genus species ------- ------ ------- ------- ------- ------- ------- ------- 9606 2759 7711 40674 9443 9604 9605 9606 9913 2759 7711 40674 91561 9895 9903 9913 376619 2 1224 1236 72273 34064 262 263 349741 2 74201 203494 48461 1647988 239934 239935 239935 2 74201 203494 48461 1647988 239934 239935 314101 2 314101 11932 10239 2732409 2732514 2169561 11632 11749 11932 1327037 10239 2731618 2731619 28883 10699 1327037 92489 2 1224 1236 91347 1903409 551 796334 1458427 2 1224 28216 80840 80864 2490452 1458425 # both node name and taxids echo 562 \ | taxonkit reformat -I 1 -t \ | csvtk -H -t sep -f 2 -s ';' -R \ | csvtk -H -t sep -f 2 -s ';' -R \ | csvtk add-header -t -n "taxid,kingdom,phylum,class,order,family,genus,species,kingdom_taxid,phylum_taxid,class_taxid,order_taxid,family_taxid,genus_taxid,species_taxid" \ | csvtk pretty -t taxid kingdom phylum class order family genus species kingdom_taxid phylum_taxid class_taxid order_taxid family_taxid genus_taxid species_taxid ----- -------- -------------- ------------------- ---------------- ------------------ ----------- ---------------- ------------- ------------ ----------- ----------- ------------ ----------- ------------- 562 Bacteria Pseudomonadota Gammaproteobacteria Enterobacterales Enterobacteriaceae Escherichia Escherichia coli 2 1224 1236 91347 543 561 562 -
Use custom symbols for unclassfied ranks (
-r/--miss-rank-repl)$ taxonkit reformat lineage.txt -r "__" | cut -f 3 Eukaryota;Chordata;Mammalia;Primates;Hominidae;Homo;Homo sapiens Eukaryota;Chordata;Mammalia;Artiodactyla;Bovidae;Bos;Bos taurus Bacteria;Proteobacteria;Gammaproteobacteria;Thiotrichales;Francisellaceae;Francisella;Francisella tularensis Bacteria;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila Bacteria;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila Bacteria;__;__;__;__;__;uncultured murine large bowel bacterium BAC 54B Viruses;Artverviricota;Revtraviricetes;Ortervirales;Retroviridae;Intracisternal A-particles;Mouse Intracisternal A-particle Viruses;Uroviricota;Caudoviricetes;Caudovirales;Siphoviridae;__;Croceibacter phage P2559Y Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacterales;Erwiniaceae;Erwinia;Erwinia oleae Bacteria;Proteobacteria;Betaproteobacteria;Burkholderiales;Comamonadaceae;Serpentinomonas;Serpentinomonas raichei $ taxonkit reformat lineage.txt -r Unassigned | cut -f 3 Eukaryota;Chordata;Mammalia;Primates;Hominidae;Homo;Homo sapiens Eukaryota;Chordata;Mammalia;Artiodactyla;Bovidae;Bos;Bos taurus Bacteria;Proteobacteria;Gammaproteobacteria;Thiotrichales;Francisellaceae;Francisella;Francisella tularensis Bacteria;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila Bacteria;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila Bacteria;Unassigned;Unassigned;Unassigned;Unassigned;Unassigned;uncultured murine large bowel bacterium BAC 54B Viruses;Artverviricota;Revtraviricetes;Ortervirales;Retroviridae;Intracisternal A-particles;Mouse Intracisternal A-particle Viruses;Uroviricota;Caudoviricetes;Caudovirales;Siphoviridae;Unassigned;Croceibacter phage P2559Y Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacterales;Erwiniaceae;Erwinia;Erwinia oleae Bacteria;Proteobacteria;Betaproteobacteria;Burkholderiales;Comamonadaceae;Serpentinomonas;Serpentinomonas raichei -
Estimate and fill missing rank with original lineage information (
-F, --fill-miss-rank, very useful for formatting input data for LEfSe). You can change the prefix "unclassified" using flag-p/--miss-rank-repl-prefix.$ cat lineage.txt \ | taxonkit reformat -F \ | csvtk -H -t cut -f 1,3 \ | csvtk -H -t sep -f 2 -s ';' -R \ | csvtk add-header -t -n taxid,kindom,phylum,class,order,family,genus,species \ | csvtk pretty -t taxid kindom phylum class order family genus species ------- --------- ---------------------------- --------------------------- --------------------------- ---------------------------- ------------------------------- ----------------------------------------------- 9606 Eukaryota Chordata Mammalia Primates Hominidae Homo Homo sapiens 9913 Eukaryota Chordata Mammalia Artiodactyla Bovidae Bos Bos taurus 376619 Bacteria Proteobacteria Gammaproteobacteria Thiotrichales Francisellaceae Francisella Francisella tularensis 349741 Bacteria Verrucomicrobia Verrucomicrobiae Verrucomicrobiales Akkermansiaceae Akkermansia Akkermansia muciniphila 239935 Bacteria Verrucomicrobia Verrucomicrobiae Verrucomicrobiales Akkermansiaceae Akkermansia Akkermansia muciniphila 314101 Bacteria unclassified Bacteria phylum unclassified Bacteria class unclassified Bacteria order unclassified Bacteria family unclassified Bacteria genus uncultured murine large bowel bacterium BAC 54B 11932 Viruses Artverviricota Revtraviricetes Ortervirales Retroviridae Intracisternal A-particles Mouse Intracisternal A-particle 1327037 Viruses Uroviricota Caudoviricetes Caudovirales Siphoviridae unclassified Siphoviridae genus Croceibacter phage P2559Y 92489 Bacteria Proteobacteria Gammaproteobacteria Enterobacterales Erwiniaceae Erwinia Erwinia oleae 1458427 Bacteria Proteobacteria Betaproteobacteria Burkholderiales Comamonadaceae Serpentinomonas Serpentinomonas raicheiDo not add prefix or suffix for estimated nodes:
$ echo 314101 | taxonkit reformat -I 1 314101 Bacteria;;;;;;uncultured murine large bowel bacterium BAC 54B $ echo 314101 | taxonkit reformat -I 1 -F -p "" -s "" 314101 Bacteria;Bacteria;Bacteria;Bacteria;Bacteria;Bacteria;uncultured murine large bowel bacterium BAC 54B -
Only some ranks.
$ cat lineage.txt \ | taxonkit reformat -F -f "{s};{p}"\ | csvtk -H -t cut -f 1,3 \ | csvtk -H -t sep -f 2 -s ';' -R \ | csvtk add-header -t -n taxid,species,phylum \ | csvtk pretty -t taxid species phylum ------- ----------------------------------------------- ---------------------------- 9606 Homo sapiens Chordata 9913 Bos taurus Chordata 376619 Francisella tularensis Proteobacteria 349741 Akkermansia muciniphila Verrucomicrobia 239935 Akkermansia muciniphila Verrucomicrobia 314101 uncultured murine large bowel bacterium BAC 54B unclassified Bacteria phylum 11932 Mouse Intracisternal A-particle Artverviricota 1327037 Croceibacter phage P2559Y Uroviricota 92489 Erwinia oleae Proteobacteria 1458427 Serpentinomonas raichei Proteobacteria -
For some taxids which rank is higher than the lowest rank in
-f/--format, use-T/--trimto avoid fill missing rank lower than current rank.$ echo -ne "2\n239934\n239935\n" \ | taxonkit lineage \ | taxonkit reformat -F \ | sed -r "s/;+$//" \ | csvtk -H -t cut -f 1,3 2 Bacteria;unclassified Bacteria phylum;unclassified Bacteria class;unclassified Bacteria order;unclassified Bacteria family;unclassified Bacteria genus;unclassified Bacteria species 239934 Bacteria;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;unclassified Akkermansia species 239935 Bacteria;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila $ echo -ne "2\n239934\n239935\n" \ | taxonkit lineage \ | taxonkit reformat -F -T \ | sed -r "s/;+$//" \ | csvtk -H -t cut -f 1,3 2 Bacteria 239934 Bacteria;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia 239935 Bacteria;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila -
Support tab in format string
$ echo 9606 \ | taxonkit lineage \ | taxonkit reformat -f "{k}\t{p}\t{c}\t{o}\t{f}\t{g}\t{s}\t{S}" \ | csvtk cut -t -f -2 9606 Eukaryota Chordata Mammalia Primates Hominidae Homo Homo sapiens -
List seven-level lineage for all TaxIds.
# replace empty taxon with "Unassigned" $ taxonkit list --ids 1 \ | taxonkit lineage \ | taxonkit reformat -r Unassigned | gzip -c > all.lineage.tsv.gz # tab-delimited seven-levels $ taxonkit list --ids 1 \ | taxonkit lineage \ | taxonkit reformat -r Unassigned -f "{k}\t{p}\t{c}\t{o}\t{f}\t{g}\t{s}" \ | csvtk cut -H -t -f -2 \ | head -n 5 \ | csvtk pretty -H -t # 8-level $ taxonkit list --ids 1 \ | taxonkit lineage \ | taxonkit reformat -r Unassigned -f "{k}\t{p}\t{c}\t{o}\t{f}\t{g}\t{s}\t{t}" \ | csvtk cut -H -t -f -2 \ | head -n 5 \ | csvtk pretty -H -t # Fill and trim $ memusg -t -s ' taxonkit list --ids 1 \ | taxonkit lineage \ | taxonkit reformat -F -T \ | sed -r "s/;+$//" \ | gzip -c > all.lineage.tsv.gz ' elapsed time: 19.930s peak rss: 6.25 GB -
From taxid to 7-ranks lineage:
$ cat taxids.txt | taxonkit lineage | taxonkit reformat # for taxonkit v0.8.0 or later versions $ cat taxids.txt | taxonkit reformat -I 1 -
Some TaxIds have the same complete lineage, empty result is returned by default. You can use the flag
-a/--output-ambiguous-resultto return one possible result. see #42$ echo -ne "2507530\n2516889\n" | taxonkit lineage --data-dir . | taxonkit reformat --data-dir . -t 19:18:29.770 [WARN] we can't distinguish the TaxIds (2507530, 2516889) for lineage: cellular organisms;Eukaryota;Opisthokonta;Fungi;Dikarya;Basidiomycota;Agaricomycotina;Agaricomycetes;Agaricomycetes incertae sedis;Russulales;Russulaceae;Russula;unclassified Russula;Russula sp. 8 KA-2019. But you can use -a/--output-ambiguous-result to return one possible result 19:18:29.770 [WARN] we can't distinguish the TaxIds (2507530, 2516889) for lineage: cellular organisms;Eukaryota;Opisthokonta;Fungi;Dikarya;Basidiomycota;Agaricomycotina;Agaricomycetes;Agaricomycetes incertae sedis;Russulales;Russulaceae;Russula;unclassified Russula;Russula sp. 8 KA-2019. But you can use -a/--output-ambiguous-result to return one possible result 2507530 cellular organisms;Eukaryota;Opisthokonta;Fungi;Dikarya;Basidiomycota;Agaricomycotina;Agaricomycetes;Agaricomycetes incertae sedis;Russulales;Russulaceae;Russula;unclassified Russula;Russula sp. 8 KA-2019 2516889 cellular organisms;Eukaryota;Opisthokonta;Fungi;Dikarya;Basidiomycota;Agaricomycotina;Agaricomycetes;Agaricomycetes incertae sedis;Russulales;Russulaceae;Russula;unclassified Russula;Russula sp. 8 KA-2019 $ echo -ne "2507530\n2516889\n" | taxonkit lineage --data-dir . | taxonkit reformat --data-dir . -t -a 2507530 cellular organisms;Eukaryota;Opisthokonta;Fungi;Dikarya;Basidiomycota;Agaricomycotina;Agaricomycetes;Agaricomycetes incertae sedis;Russulales;Russulaceae;Russula;unclassified Russula;Russula sp. 8 KA-2019 Eukaryota;Basidiomycota;Agaricomycetes;Russulales;Russulaceae;Russula;Russula sp. 8 KA-2019 2759;5204;155619;452342;5401;5402;2507530 2516889 cellular organisms;Eukaryota;Opisthokonta;Fungi;Dikarya;Basidiomycota;Agaricomycotina;Agaricomycetes;Agaricomycetes incertae sedis;Russulales;Russulaceae;Russula;unclassified Russula;Russula sp. 8 KA-2019 Eukaryota;Basidiomycota;Agaricomycetes;Russulales;Russulaceae;Russula;Russula sp. 8 KA-2019 2759;5204;155619;452342;5401;5402;2507530
reformat2Link
Usage
Reformat lineage in chosen ranks, allowing more ranks than 'reformat'
Input:
- List of TaxIds, one record per line.
- Or tab-delimited format.
Please specify the TaxId field with flag -I/--taxid-field (default 1)
- Supporting (gzipped) file or STDIN.
Output:
1. Input line data.
2. Reformated lineage.
3. (Optional) TaxIds taxons in the lineage (-t/--show-lineage-taxids)
Output format:
1. It can contain some escape characters like "\t".
2. You can use "|" to set multiple ranks, and the first valid one will be outputted.
This is useful for a rank with different rank names, especially since NCBI
made big changes to some ranks in March 2025:
- "Domain" replaces "superkingdom" for Archaea, Bacteria, and Eukaryota
- "Acellular root" replaces "superkingdom" for Viruses
- Six viral groups are designated with the new rank "realm", the equivalent of "domain"
So, we can use "{domain|acellular root|superkingdom}" to handle all these cases
and keep compatible with old taxonomy data.
$ echo -ne "Eukaryota\nBacteria\nViruses\n" \
| taxonkit name2taxid -s -r \
| taxonkit reformat2 -I 2 -f "{domain|acellular root|superkingdom}" \
| csvtk add-header -Ht -n name,taxid,rank,kingdom/domain \
| csvtk pretty -t
name taxid rank kingdom/domain
--------- ----- -------------- --------------
Eukaryota 2759 domain Eukaryota
Bacteria 2 domain Bacteria
Viruses 10239 acellular root Viruses
Another example is for subspecies nodes, the rank might be "subpecies", "strain", or "no rank".
For example,
$ echo -ne "562\n83333\n2697049\n" \
| taxonkit lineage -L -r \
| taxonkit reformat2 -f "{species};{strain|subspecies|no rank}"
562 species Escherichia coli;
83333 strain Escherichia coli;Escherichia coli K-12
2697049 no rank Severe acute respiratory syndrome-related coronavirus;Severe acute respiratory syndrome coronavirus 2
Differences from 'taxonkit reformat':
- [input] only accept TaxIDs
- [format] accept more rank place holders, not just the seven canonical ones.
- [format] use the full name of ranks, such as "{species}", rather than "{s}"
- [format] support multiple ranks in one place holder, such as "{subspecies|strain}"
- do not automatically add prefixes, but you can simply set them in the format
Usage:
taxonkit reformat2 [flags]
Flags:
-f, --format string output format, placeholders of rank are needed (default
"{domain|acellular
root|superkingdom};{phylum};{class};{order};{family};{genus};{species}")
-h, --help help for reformat2
-r, --miss-rank-repl string replacement string for missing rank
-R, --miss-taxid-repl string replacement string for missing taxid
-B, --no-ranks strings rank names of no-rank. A lineage might have many "no rank" ranks, we
only keep the last one below known ranks (default [no rank,clade])
-t, --show-lineage-taxids show corresponding taxids of reformated lineage
-I, --taxid-field int field index of taxid. input data should be tab-separated. it overrides
-i/--lineage-field (default 1)
-T, --trim do not replace missing ranks lower than the rank of the current node
Examples
-
Default format
$ echo 562 | taxonkit reformat2 562 Bacteria;Pseudomonadota;Gammaproteobacteria;Enterobacterales;Enterobacteriaceae;Escherichia;Escherichia coli -
Change the format
$ echo 562 | taxonkit reformat2 -f "g__{genus}\ts__{species}" 562 g__Escherichia s__Escherichia coli -
Subspecies
$ echo 511145 | taxonkit lineage 511145 cellular organisms;Bacteria;Pseudomonadota;Gammaproteobacteria;Enterobacterales;Enterobacteriaceae;Escherichia;Escherichia coli;Escherichia coli K-12;Escherichia coli str. K-12 substr. MG1655 $ echo 511145 | taxonkit reformat -I 1 -f "{d};{p};{c};{o};{f};{g};{s};{t}" 511145 Bacteria;Pseudomonadota;Gammaproteobacteria;Enterobacterales;Enterobacteriaceae;Escherichia;Escherichia coli;Escherichia coli K-12 $ echo 511145 | taxonkit reformat2 -I 1 -f "{domain|acellular root|superkingdom};{phylum};{class};{order};{family};{genus};{species};{subspecies|strain|no rank}" 511145 Bacteria;Pseudomonadota;Gammaproteobacteria;Enterobacterales;Enterobacteriaceae;Escherichia;Escherichia coli;Escherichia coli K-12 -
Trim
$ echo 561 | taxonkit reformat2 -r unknown 561 Bacteria;Pseudomonadota;Gammaproteobacteria;Enterobacterales;Enterobacteriaceae;Escherichia;unknown $ echo 561 | taxonkit reformat2 -r unknown -T 561 Bacteria;Pseudomonadota;Gammaproteobacteria;Enterobacterales;Enterobacteriaceae;Escherichia; # ----------------------------------------------------------- # another example where the order rank is missing $ echo 102403 | taxonkit reformat2 -I 1 -r "0" -f "{domain|acellular root|superkingdom};{phylum};{class};{order};{family};{genus};{species}" 102403 Eukaryota;Mollusca;Bivalvia;0;Poromyidae;Tropidomya;0 $ echo 102403 | taxonkit reformat2 -I 1 -r "0" -f "{domain|acellular root|superkingdom};{phylum};{class};{order};{family};{genus};{species}" -T 102403 Eukaryota;Mollusca;Bivalvia;0;Poromyidae;Tropidomya; # and now, the lowest rank in the output format is order, but the tailing "0" is not trimmed. $ echo 102403 | taxonkit reformat2 -I 1 -r "0" -f "{domain|acellular root|superkingdom};{phylum};{class};{order}" 102403 Eukaryota;Mollusca;Bivalvia;0 $ echo 102403 | taxonkit reformat2 -I 1 -r "0" -f "{domain|acellular root|superkingdom};{phylum};{class};{order}" -T 102403 Eukaryota;Mollusca;Bivalvia;0
name2taxidLink
Usage
Convert taxon names to TaxIds
Attention:
1. Some TaxIds share the same names, e.g, Drosophila.
These input lines are duplicated with multiple TaxIds.
$ echo Drosophila | taxonkit name2taxid | taxonkit lineage -i 2 -r -L
Drosophila 7215 genus
Drosophila 32281 subgenus
Drosophila 2081351 genus
Usage:
taxonkit name2taxid [flags]
Flags:
-h, --help help for name2taxid
-i, --name-field int field index of name. data should be tab-separated (default 1)
-s, --sci-name only searching scientific names
-r, --show-rank show rank
Examples
Example data
$ cat names.txt
Homo sapiens
Akkermansia muciniphila ATCC BAA-835
Akkermansia muciniphila
Mouse Intracisternal A-particle
Wei Shen
uncultured murine large bowel bacterium BAC 54B
Croceibacter phage P2559Y
-
Default.
# taxonkit name2taxid names.txt $ cat names.txt | taxonkit name2taxid | csvtk pretty -H -t Homo sapiens 9606 Akkermansia muciniphila ATCC BAA-835 349741 Akkermansia muciniphila 239935 Mouse Intracisternal A-particle 11932 Wei Shen uncultured murine large bowel bacterium BAC 54B 314101 Croceibacter phage P2559Y 1327037 -
Show rank.
$ cat names.txt | taxonkit name2taxid --show-rank | csvtk pretty -H -t Homo sapiens 9606 species Akkermansia muciniphila ATCC BAA-835 349741 strain Akkermansia muciniphila 239935 species Mouse Intracisternal A-particle 11932 species Wei Shen uncultured murine large bowel bacterium BAC 54B 314101 species Croceibacter phage P2559Y 1327037 species -
From name to lineage.
$ cat names.txt | taxonkit name2taxid | taxonkit lineage --taxid-field 2 Homo sapiens 9606 cellular organisms;Eukaryota;Opisthokonta;Metazoa;Eumetazoa;Bilateria;Deuterostomia;Chordata;Craniata;Vertebrata;Gnathostomata;Teleostomi;Euteleostomi;Sarcopterygii;Dipnotetrapodomorpha;Tetrapoda;Amniota;Mammalia;Theria;Eutheria;Boreoeutheria;Euarchontoglires;Primates;Haplorrhini;Simiiformes;Catarrhini;Hominoidea;Hominidae;Homininae;Homo;Homo sapiens Akkermansia muciniphila ATCC BAA-835 349741 cellular organisms;Bacteria;PVC group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila;Akkermansia muciniphila ATCC BAA-835 Akkermansia muciniphila 239935 cellular organisms;Bacteria;PVC group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila Mouse Intracisternal A-particle 11932 Viruses;Ortervirales;Retroviridae;unclassified Retroviridae;Intracisternal A-particles;Mouse Intracisternal A-particle Wei Shen uncultured murine large bowel bacterium BAC 54B 314101 cellular organisms;Bacteria;environmental samples;uncultured murine large bowel bacterium BAC 54B Croceibacter phage P2559Y 1327037 Viruses;Caudovirales;Siphoviridae;unclassified Siphoviridae;Croceibacter phage P2559Y -
Convert old names to new names.
$ echo Lactobacillus fermentum | taxonkit name2taxid | taxonkit lineage -i 2 -n | cut -f 1,2,4 Lactobacillus fermentum 1613 Limosilactobacillus fermentum -
Some TaxIds share the same scientific names, e.g, Drosophila.
$ echo Drosophila \ | taxonkit name2taxid \ | taxonkit lineage -i 2 -r \ | taxonkit reformat -i 3 \ | csvtk cut -H -t -f 1,2,4,5 \ | csvtk pretty -H -t Drosophila 7215 genus Eukaryota;Arthropoda;Insecta;Diptera;Drosophilidae;Drosophila; Drosophila 32281 subgenus Eukaryota;Arthropoda;Insecta;Diptera;Drosophilidae;Drosophila; Drosophila 2081351 genus Eukaryota;Basidiomycota;Agaricomycetes;Agaricales;Psathyrellaceae;Drosophila;
filterLink
Usage
Filter TaxIds by taxonomic rank range
Attention:
1. Flag -L/--lower-than and -H/--higher-than are exclusive, and can be
used along with -E/--equal-to which values can be different.
2. A list of pre-ordered ranks is in ~/.taxonkit/ranks.txt, you can use
your list by -r/--rank-file, the format specification is below.
3. All ranks in taxonomy database should be defined in rank file.
4. Ranks can be removed with black list via -B/--black-list.
5. TaxIDs with no rank (those starting with ! in the rank file) are kept
by default! They can be optionally discarded by -N/--discard-noranks.
One exception: -N/--discard-noranks is switched on automatically
when only -E/--equal-to is given and the value is not one of ranks
without order ("no rank", "clade").
6. [Recommended] When filtering with -L/--lower-than, you can use
-n/--save-predictable-norank to save some special ranks without order,
where rank of the closest higher node is still lower than rank cutoff.
Rank file:
1. Blank lines or lines starting with "#" are ignored.
2. Ranks are in decending order and case ignored.
3. Ranks with same order should be in one line separated with comma (",", no space).
4. Ranks without order should be assigned a prefix symbol "!" for each rank.
Usage:
taxonkit filter [flags]
Flags:
-B, --black-list strings black list of ranks to discard, e.g., '-B "no rank" -B "clade"
-N, --discard-noranks discard all ranks without order, type "taxonkit filter --help" for details
-R, --discard-root discard root taxid, defined by --root-taxid
-E, --equal-to strings output TaxIds with rank equal to some ranks, multiple values can be
separated with comma "," (e.g., -E "genus,species"), or give multiple
times (e.g., -E genus -E species)
-h, --help help for filter
-H, --higher-than string output TaxIds with rank higher than a rank, exclusive with --lower-than
--list-order list user defined ranks in order, from "$HOME/.taxonkit/ranks.txt"
--list-ranks list ordered ranks in taxonomy database, sorted in user defined order
-L, --lower-than string output TaxIds with rank lower than a rank, exclusive with --higher-than
-r, --rank-file string user-defined ordered taxonomic ranks, type "taxonkit filter --help"
for details
--root-taxid uint32 root taxid (default 1)
-n, --save-predictable-norank do not discard some special ranks without order when using -L, where
rank of the closest higher node is still lower than rank cutoff
-i, --taxid-field int field index of taxid. input data should be tab-separated (default 1)
Examples
-
Example data
$ echo 349741 | taxonkit lineage -t | cut -f 3 | sed 's/;/\n/g' > taxids2.txt $ cat taxids2.txt 131567 2 1783257 74201 203494 48461 1647988 239934 239935 349741 $ cat taxids2.txt | taxonkit lineage -r | csvtk -Ht cut -f 1,3,2 | csvtk pretty -H -t 131567 cellular root cellular organisms 2 domain cellular organisms;Bacteria 1783257 clade cellular organisms;Bacteria;Pseudomonadati;PVC group 74201 phylum cellular organisms;Bacteria;Pseudomonadati;PVC group;Verrucomicrobiota 203494 class cellular organisms;Bacteria;Pseudomonadati;PVC group;Verrucomicrobiota;Verrucomicrobiia 48461 order cellular organisms;Bacteria;Pseudomonadati;PVC group;Verrucomicrobiota;Verrucomicrobiia;Verrucomicrobiales 1647988 family cellular organisms;Bacteria;Pseudomonadati;PVC group;Verrucomicrobiota;Verrucomicrobiia;Verrucomicrobiales;Akkermansiaceae 239934 genus cellular organisms;Bacteria;Pseudomonadati;PVC group;Verrucomicrobiota;Verrucomicrobiia;Verrucomicrobiales;Akkermansiaceae;Akkermansia 239935 species cellular organisms;Bacteria;Pseudomonadati;PVC group;Verrucomicrobiota;Verrucomicrobiia;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila 349741 strain cellular organisms;Bacteria;Pseudomonadati;PVC group;Verrucomicrobiota;Verrucomicrobiia;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila;Akkermansia muciniphila ATCC BAA-835 -
Equal to certain rank(s) (
-E/--equal-to)$ cat taxids2.txt \ | taxonkit filter -E Phylum -E Class -N \ | taxonkit lineage -r \ | csvtk -Ht cut -f 1,3,2 \ | csvtk pretty -H -t 74201 phylum cellular organisms;Bacteria;PVC group;Verrucomicrobia 203494 class cellular organisms;Bacteria;PVC group;Verrucomicrobia;Verrucomicrobiae -
Lower than a rank (
-L/--lower-than)$ cat taxids2.txt \ | taxonkit filter -L genus -N \ | taxonkit lineage -r -n -L \ | csvtk -Ht cut -f 1,3,2 \ | csvtk pretty -H -t 239935 species Akkermansia muciniphila 349741 strain Akkermansia muciniphila ATCC BAA-835 -
Higher than a rank (
-H/--higher-than)$ cat taxids2.txt \ | taxonkit filter -H phylum -N \ | taxonkit lineage -r -n -L \ | csvtk -Ht cut -f 1,3,2 \ | csvtk pretty -H -t 131567 cellular root cellular organisms 2 domain Bacteria -
TaxIDs with no rank are kept by default!!! "no rank" and "clade" have no rank and can be filter out via
-N/--discard-noranks. Futher ranks can be removed with black list via-B/--black-list.# 562 is the TaxId of Escherichia coli $ taxonkit list --ids 562 \ | taxonkit filter -L species \ | taxonkit lineage -r -n -L \ | csvtk cut -Ht -f 1,3,2 \ | csvtk freq -Ht -f 2 -nr \ | csvtk pretty -H -t strain 2940 no rank 486 serotype 176 serogroup 110 isolate 1 subspecies 1 $ taxonkit list --ids 562 \ | taxonkit filter -L species -N -B strain \ | taxonkit lineage -r -n -L \ | csvtk cut -Ht -f 1,3,2 \ | csvtk freq -Ht -f 2 -nr \ | csvtk pretty -H -t serotype 176 serogroup 110 isolate 1 subspecies 1 -
Combine of
-L/-Hwith-E.$ cat taxids2.txt \ | taxonkit filter -L genus -E genus -N \ | taxonkit lineage -r -n -L \ | csvtk cut -Ht -f 1,3,2 \ | csvtk pretty -H -t 239934 genus Akkermansia 239935 species Akkermansia muciniphila 349741 strain Akkermansia muciniphila ATCC BAA-835 -
Special cases of "no rank". (
-n/--save-predictable-norank). When filtering with-L/--lower-than, you can use-n/--save-predictable-norankto save some special ranks without order, where rank of the closest higher node is still lower than rank cutoff.$ echo -ne "2605619\n1327037\n" \ | taxonkit lineage -t \ | csvtk cut -Ht -f 3 \ | csvtk unfold -Ht -f 1 -s ";" \ | taxonkit lineage -r -n -L \ | csvtk cut -Ht -f 1,3,2 \ | csvtk pretty -H -t 131567 cellular root cellular organisms 2 domain Bacteria 3379134 kingdom Pseudomonadati 1224 phylum Pseudomonadota 1236 class Gammaproteobacteria 91347 order Enterobacterales 543 family Enterobacteriaceae 561 genus Escherichia 562 species Escherichia coli 2605619 no rank Escherichia coli O16:H48 10239 acellular root Viruses 2731341 realm Duplodnaviria 2731360 kingdom Heunggongvirae 2731618 phylum Uroviricota 2731619 class Caudoviricetes 2788787 no rank unclassified Caudoviricetes 1327037 species Croceibacter phage P2559Y # save taxids $ echo -ne "2605619\n1327037\n" \ | taxonkit lineage -t \ | csvtk cut -Ht -f 3 \ | csvtk unfold -Ht -f 1 -s ";" \ | tee taxids4.txt 131567 2 1224 1236 91347 543 561 562 2605619 10239 2731341 2731360 2731618 2731619 28883 10699 196894 1327037Now, filter nodes of rank <= species.
$ cat taxids4.txt \ | taxonkit filter -L species -E species -N -n \ | taxonkit lineage -r -n -L \ | csvtk cut -Ht -f 1,3,2 \ | csvtk pretty -H -t 562 species Escherichia coli 2605619 no rank Escherichia coli O16:H48 1327037 species Croceibacter phage P2559YNote that 2605619 (no rank) is saved because its parent node 562 is <= species.
lcaLink
Usage
Compute lowest common ancestor (LCA) for TaxIds
Attention:
1. This command computes LCA TaxId for a list of TaxIds
in a field ("-i/--taxids-field) of tab-delimited file or STDIN.
2. TaxIDs should have the same separator ("-s/--separator"),
single charactor separator is prefered.
3. Empty lines or lines without valid TaxIds in the field are omitted.
4. If some TaxIds are not found in database, it returns 0.
Examples:
$ echo 239934, 239935, 349741 | taxonkit lca -s ", "
239934, 239935, 349741 239934
$ time echo 239934 239935 349741 9606 | taxonkit lca
239934 239935 349741 9606 131567
Usage:
taxonkit lca [flags]
Flags:
-b, --buffer-size string size of line buffer, supported unit: K, M, G. You need to increase the
value when "bufio.Scanner: token too long" error occured (default "1M")
-h, --help help for lca
--separater string separater for TaxIds. This flag is same to --separator. (default " ")
-s, --separator string separator for TaxIds (default " ")
-D, --skip-deleted skip deleted TaxIds and compute with left ones
-U, --skip-unfound skip unfound TaxIds and compute with left ones
-i, --taxids-field int field index of TaxIds. Input data should be tab-separated (default 1)
Examples:
-
Example data
$ taxonkit list --ids 9605 -nr --indent " " 9605 [genus] Homo 9606 [species] Homo sapiens 63221 [subspecies] Homo sapiens neanderthalensis 741158 [subspecies] Homo sapiens subsp. 'Denisova' 1425170 [species] Homo heidelbergensis 2665952 [no rank] environmental samples 2665953 [species] Homo sapiens environmental sample -
Simple one
$ echo 63221 2665953 | taxonkit lca 63221 2665953 9605 -
Custom field (
-i/--taxids-field) and separater (-s/--separator).$ echo -ne "a\t63221,2665953\nb\t63221, 741158\n" a 63221,2665953 b 63221, 741158 $ echo -ne "a\t63221,2665953\nb\t63221, 741158\n" \ | taxonkit lca -i 2 -s "," a 63221,2665953 9605 b 63221, 741158 9606 -
Merged TaxIds.
# merged $ echo 92487 92488 92489 | taxonkit lca 10:08:26.578 [WARN] taxid 92489 was merged into 796334 92487 92488 92489 1236 -
Deleted TaxIds, you can ommit theses and continue compute with left onces with (
-D/--skip-deleted).$ echo 1 2 3 | taxonkit lca 10:30:17.678 [WARN] taxid 3 not found 1 2 3 0 $ time echo 1 2 3 | taxonkit lca -D 10:29:31.828 [WARN] taxid 3 was deleted 1 2 3 1 -
TaxIDs not found in database, you can ommit theses and continue compute with left onces with (
-U/--skip-unfound).$ echo 61021 61022 11111111 | taxonkit lca 10:31:44.929 [WARN] taxid 11111111 not found 61021 61022 11111111 0 $ echo 61021 61022 11111111 | taxonkit lca -U 10:32:02.772 [WARN] taxid 11111111 not found 61021 61022 11111111 2628496
taxid-changelogLink
Usage
Create TaxId changelog from dump archives
Attention:
1. This command was originally designed for NCBI taxonomy, where the the TaxIds are stable.
2. For other taxonomic data created by "taxonkit create-taxdump", e.g., GTDB-taxdump,
some change events might be wrong, because
a) There would be dramatic changes between the two versions.
b) Different taxons in multiple versions might have the same TaxIds, because we only
check and eliminate taxid collision within a single version.
So a single version of taxonomic data created by "taxonkit create-taxdump" has no problem,
it's just the changelog might not be perfect.
Steps:
# dependencies:
# rush - https://github.com/shenwei356/rush/
mkdir -p archive; cd archive;
# --------- download ---------
# option 1
# for fast network connection
wget ftp://ftp.ncbi.nlm.nih.gov/pub/taxonomy/taxdump_archive/taxdmp*.zip
# option 2
# for slow network connection
url=https://ftp.ncbi.nlm.nih.gov/pub/taxonomy/taxdump_archive/
wget $url -O - -o /dev/null \
| grep taxdmp | perl -ne '/(taxdmp_.+?.zip)/; print "$1\n";' \
| rush -j 2 -v url=$url 'axel -n 5 {url}/{}' \
--immediate-output -c -C download.rush
# --------- unzip ---------
ls taxdmp*.zip | rush -j 1 'unzip {} names.dmp nodes.dmp merged.dmp delnodes.dmp -d {@_(.+)\.}'
# optionally compress .dmp files with pigz, for saving disk space
fd .dmp$ | rush -j 4 'pigz {}'
# --------- create log ---------
cd ..
taxonkit taxid-changelog -i archive -o taxid-changelog.csv.gz --verbose
Output format (CSV):
# fields comments
taxid # taxid
version # version / time of archive, e.g, 2019-07-01
change # change, values:
# NEW newly added
# REUSE_DEL deleted taxids being reused
# REUSE_MER merged taxids being reused
# DELETE deleted
# MERGE merged into another taxid
# ABSORB other taxids merged into this one
# CHANGE_NAME scientific name changed
# CHANGE_RANK rank changed
# CHANGE_LIN_LIN lineage taxids remain but lineage remain
# CHANGE_LIN_TAX lineage taxids changed
# CHANGE_LIN_LEN lineage length changed
change-value # variable values for changes:
# 1) new taxid for MERGE
# 2) merged taxids for ABSORB
# 3) empty for others
name # scientific name
rank # rank
lineage # complete lineage of the taxid
lineage-taxids # taxids of the lineage
# you can use csvtk to investigate them. e.g.,
csvtk grep -f taxid -p 1390515 taxid-changelog.csv.gz
Usage:
taxonkit taxid-changelog [flags]
Flags:
-i, --archive string directory containing uncompressed dumped archives
-h, --help help for taxid-changelog
-
Example 1 (E.coli with taxid
562)$ pigz -cd taxid-changelog.csv.gz \ | csvtk grep -f taxid -p 562 \ | csvtk pretty taxid version change change-value name rank lineage lineage-taxids 562 2014-08-01 NEW Escherichia coli species cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacteriales;Enterobacteriaceae;Escherichia;Escherichia coli 131567;2;1224;1236;91347;543;561;562 562 2014-08-01 ABSORB 662101;662104 Escherichia coli species cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacteriales;Enterobacteriaceae;Escherichia;Escherichia coli 131567;2;1224;1236;91347;543;561;562 562 2015-11-01 ABSORB 1637691 Escherichia coli species cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacteriales;Enterobacteriaceae;Escherichia;Escherichia coli 131567;2;1224;1236;91347;543;561;562 562 2016-10-01 CHANGE_LIN_LIN Escherichia coli species cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacterales;Enterobacteriaceae;Escherichia;Escherichia coli 131567;2;1224;1236;91347;543;561;562 562 2018-06-01 ABSORB 469598 Escherichia coli species cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacterales;Enterobacteriaceae;Escherichia;Escherichia coli 131567;2;1224;1236;91347;543;561;562 # merged taxids $ pigz -cd taxid-changelog.csv.gz \ | csvtk grep -f taxid -p 662101,662104,1637691,469598 \ | csvtk pretty taxid version change change-value name rank lineage lineage-taxids 469598 2014-08-01 NEW Escherichia sp. 3_2_53FAA species cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacteriales;Enterobacteriaceae;Escherichia;Escherichia sp. 3_2_53FAA 131567;2;1224;1236;91347;543;561;469598 469598 2016-10-01 CHANGE_LIN_LIN Escherichia sp. 3_2_53FAA species cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacterales;Enterobacteriaceae;Escherichia;Escherichia sp. 3_2_53FAA 131567;2;1224;1236;91347;543;561;469598 469598 2018-06-01 MERGE 562 Escherichia sp. 3_2_53FAA species cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacterales;Enterobacteriaceae;Escherichia;Escherichia sp. 3_2_53FAA 131567;2;1224;1236;91347;543;561;469598 662101 2014-08-01 MERGE 562 662104 2014-08-01 MERGE 562 1637691 2015-04-01 DELETE 1637691 2015-05-01 REUSE_DEL Escherichia sp. MAR species cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacteriales;Enterobacteriaceae;Escherichia;Escherichia sp. MAR 131567;2;1224;1236;91347;543;561;1637691 1637691 2015-11-01 MERGE 562 Escherichia sp. MAR species cellular organisms;Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacteriales;Enterobacteriaceae;Escherichia;Escherichia sp. MAR 131567;2;1224;1236;91347;543;561;1637691 -
Example 2 (SARS-CoV-2).
$ time pigz -cd taxid-changelog.csv.gz \ | csvtk grep -f taxid -p 2697049 \ | csvtk pretty taxid version change change-value name rank lineage lineage-taxids 2697049 2020-02-01 NEW Wuhan seafood market pneumonia virus species Viruses;Riboviria;Nidovirales;Cornidovirineae;Coronaviridae;Orthocoronavirinae;Betacoronavirus;unclassified Betacoronavirus;Wuhan seafood market pneumonia virus 10239;2559587;76804;2499399;11118;2501931;694002;696098;2697049 2697049 2020-03-01 CHANGE_NAME Severe acute respiratory syndrome coronavirus 2 no rank Viruses;Riboviria;Nidovirales;Cornidovirineae;Coronaviridae;Orthocoronavirinae;Betacoronavirus;Sarbecovirus;Severe acute respiratory syndrome-related coronavirus;Severe acute respiratory syndrome coronavirus 2 10239;2559587;76804;2499399;11118;2501931;694002;2509511;694009;2697049 2697049 2020-03-01 CHANGE_RANK Severe acute respiratory syndrome coronavirus 2 no rank Viruses;Riboviria;Nidovirales;Cornidovirineae;Coronaviridae;Orthocoronavirinae;Betacoronavirus;Sarbecovirus;Severe acute respiratory syndrome-related coronavirus;Severe acute respiratory syndrome coronavirus 2 10239;2559587;76804;2499399;11118;2501931;694002;2509511;694009;2697049 2697049 2020-03-01 CHANGE_LIN_LEN Severe acute respiratory syndrome coronavirus 2 no rank Viruses;Riboviria;Nidovirales;Cornidovirineae;Coronaviridae;Orthocoronavirinae;Betacoronavirus;Sarbecovirus;Severe acute respiratory syndrome-related coronavirus;Severe acute respiratory syndrome coronavirus 2 10239;2559587;76804;2499399;11118;2501931;694002;2509511;694009;2697049 2697049 2020-06-01 CHANGE_LIN_LEN Severe acute respiratory syndrome coronavirus 2 no rank Viruses;Riboviria;Orthornavirae;Pisuviricota;Pisoniviricetes;Nidovirales;Cornidovirineae;Coronaviridae;Orthocoronavirinae;Betacoronavirus;Sarbecovirus;Severe acute respiratory syndrome-related coronavirus;Severe acute respiratory syndrome coronavirus 2 10239;2559587;2732396;2732408;2732506;76804;2499399;11118;2501931;694002;2509511;694009;2697049 2697049 2020-07-01 CHANGE_RANK Severe acute respiratory syndrome coronavirus 2 isolate Viruses;Riboviria;Orthornavirae;Pisuviricota;Pisoniviricetes;Nidovirales;Cornidovirineae;Coronaviridae;Orthocoronavirinae;Betacoronavirus;Sarbecovirus;Severe acute respiratory syndrome-related coronavirus;Severe acute respiratory syndrome coronavirus 2 10239;2559587;2732396;2732408;2732506;76804;2499399;11118;2501931;694002;2509511;694009;2697049 2697049 2020-08-01 CHANGE_RANK Severe acute respiratory syndrome coronavirus 2 no rank Viruses;Riboviria;Orthornavirae;Pisuviricota;Pisoniviricetes;Nidovirales;Cornidovirineae;Coronaviridae;Orthocoronavirinae;Betacoronavirus;Sarbecovirus;Severe acute respiratory syndrome-related coronavirus;Severe acute respiratory syndrome coronavirus 2 10239;2559587;2732396;2732408;2732506;76804;2499399;11118;2501931;694002;2509511;694009;2697049 real 0m7.644s user 0m16.749s sys 0m3.985s -
Example 3 (All subspecies and strain in Akkermansia muciniphila 239935)
# species in Akkermansia $ taxonkit list --show-rank --show-name --indent " " --ids 239935 239935 [species] Akkermansia muciniphila 349741 [strain] Akkermansia muciniphila ATCC BAA-835 # check them all $ pigz -cd taxid-changelog.csv.gz \ | csvtk grep -f taxid -P <(taxonkit list --indent "" --ids 239935) \ | csvtk pretty lineage-taxids taxid version change change-value name rank lineage lineage-taxids 239935 2014-08-01 NEW Akkermansia muciniphila species cellular organisms;Bacteria;Chlamydiae/Verrucomicrobia group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Verrucomicrobiaceae;Akkermansia;Akkermansia muciniphila 131567;2;51290;74201;203494;48461;203557;239934;239935 239935 2015-05-01 CHANGE_LIN_TAX Akkermansia muciniphila species cellular organisms;Bacteria;Chlamydiae/Verrucomicrobia group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila 131567;2;51290;74201;203494;48461;1647988;239934;239935 239935 2016-03-01 CHANGE_LIN_TAX Akkermansia muciniphila species cellular organisms;Bacteria;PVC group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila 131567;2;1783257;74201;203494;48461;1647988;239934;239935 239935 2016-05-01 ABSORB 1834199 Akkermansia muciniphila species cellular organisms;Bacteria;PVC group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila 131567;2;1783257;74201;203494;48461;1647988;239934;239935 349741 2014-08-01 NEW Akkermansia muciniphila ATCC BAA-835 no rank cellular organisms;Bacteria;Chlamydiae/Verrucomicrobia group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Verrucomicrobiaceae;Akkermansia;Akkermansia muciniphila;Akkermansia muciniphila ATCC BAA-835 131567;2;51290;74201;203494;48461;203557;239934;239935;349741 349741 2015-05-01 CHANGE_LIN_TAX Akkermansia muciniphila ATCC BAA-835 no rank cellular organisms;Bacteria;Chlamydiae/Verrucomicrobia group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila;Akkermansia muciniphila ATCC BAA-835 131567;2;51290;74201;203494;48461;1647988;239934;239935;349741 349741 2016-03-01 CHANGE_LIN_TAX Akkermansia muciniphila ATCC BAA-835 no rank cellular organisms;Bacteria;PVC group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila;Akkermansia muciniphila ATCC BAA-835 131567;2;1783257;74201;203494;48461;1647988;239934;239935;349741 349741 2020-07-01 CHANGE_RANK Akkermansia muciniphila ATCC BAA-835 strain cellular organisms;Bacteria;PVC group;Verrucomicrobia;Verrucomicrobiae;Verrucomicrobiales;Akkermansiaceae;Akkermansia;Akkermansia muciniphila;Akkermansia muciniphila ATCC BAA-835 131567;2;1783257;74201;203494;48461;1647988;239934;239935;349741
create-taxdumpLink
Usage
Create NCBI-style taxdump files for custom taxonomy, e.g., GTDB and ICTV
Input format:
0. For GTDB taxonomy file, just use --gtdb.
We use the numeric assembly accession as the taxon at subspecies rank.
(without the prefix GCA_ and GCF_, and version number).
1. The input file should be tab-delimited, at least one column is needed.
2. Ranks can be given either via the first row or the flag --rank-names.
3. The column containing the genome/assembly accession is recommended to
generate TaxId mapping file (taxid.map, id -> taxid).
-A/--field-accession, field contaning genome/assembly accession
--field-accession-re, regular expression to extract the accession
Note that mutiple TaxIds pointing to the same accession are listed as
comma-seperated integers.
Attention:
1. Duplicated taxon names wit different ranks are allowed since v0.16.0, since
the rank and taxon name are contatenated for generating the TaxId.
2. The generated TaxIds are not consecutive numbers, however some tools like MMSeqs2
required this, you can use the script below for convertion:
https://github.com/apcamargo/ictv-mmseqs2-protein-database/blob/master/scripts/fix_taxdump.py
3. We only check and eliminate taxid collision within a single version of taxonomy data.
Therefore, if you create taxid-changelog with "taxid-changelog", different taxons
in multiple versions might have the same TaxIds and some change events might be wrong.
So a single version of taxonomic data created by "taxonkit create-taxdump" has no problem,
it's just the changelog might not be perfect.
Usage:
taxonkit create-taxdump [flags]
Flags:
-A, --field-accession int field index of assembly accession (genome ID), for outputting
taxid.map
-S, --field-accession-as-subspecies treate the accession as subspecies rank
--field-accession-re string regular expression to extract assembly accession (default "^(.+)$")
--force overwrite existing output directory
--gtdb input files are GTDB taxonomy file
--gtdb-re-subs string regular expression to extract assembly accession as the
subspecies (default "^\\w\\w_GC[AF]_(.+)\\.\\d+$")
-h, --help help for create-taxdump
--line-chunk-size int number of lines to process for each thread, and 4 threads is
fast enough. (default 5000)
--null strings null value of taxa (default [,NULL,NA])
-x, --old-taxdump-dir string taxdump directory of the previous version, for generating
merged.dmp and delnodes.dmp
-O, --out-dir string output directory
-R, --rank-names strings names of all ranks, leave it empty to use the (lowercase) first
row of input as rank names
Examples:
-
GTDB. See more: https://github.com/shenwei356/gtdb-taxdump
$ taxonkit create-taxdump --gtdb ar53_taxonomy_r207.tsv.gz bac120_taxonomy_r207.tsv.gz --out-dir taxdump 16:42:35.213 [INFO] 317542 records saved to taxdump/taxid.map 16:42:35.460 [INFO] 401815 records saved to taxdump/nodes.dmp 16:42:35.611 [INFO] 401815 records saved to taxdump/names.dmp 16:42:35.611 [INFO] 0 records saved to taxdump/merged.dmp 16:42:35.611 [INFO] 0 records saved to taxdump/delnodes.dmp -
ICTV, See more: https://github.com/shenwei356/ictv-taxdump
-
MGV. Only Order, Family, Genus information are available.
$ cat mgv_contig_info.tsv \ | csvtk cut -t -f ictv_order,ictv_family,ictv_genus,votu_id,contig_id \ | sed 1d \ > mgv.tsv $ taxonkit create-taxdump mgv.tsv --out-dir mgv --force -A 5 -R order,family,genus,species 23:33:18.098 [INFO] 189680 records saved to mgv/taxid.map 23:33:18.131 [INFO] 58102 records saved to mgv/nodes.dmp 23:33:18.150 [INFO] 58102 records saved to mgv/names.dmp 23:33:18.150 [INFO] 0 records saved to mgv/merged.dmp 23:33:18.150 [INFO] 0 records saved to mgv/delnodes.dmp $ head -n 5 mgv/taxid.map MGV-GENOME-0364295 677052301 MGV-GENOME-0364296 677052301 MGV-GENOME-0364303 1414406025 MGV-GENOME-0364311 1849074420 MGV-GENOME-0364312 2074846424 $ echo 677052301 | taxonkit lineage --data-dir mgv/ 677052301 Caudovirales;crAss-phage;OTU-61123 $ echo 677052301 | taxonkit reformat --data-dir mgv/ -I 1 -P 677052301 k__;p__;c__;o__Caudovirales;f__crAss-phage;g__;s__OTU-61123 $ grep MGV-GENOME-0364295 mgv.tsv Caudovirales crAss-phage NULL OTU-61123 MGV-GENOME-0364295 -
Custom lineages with the first row as rank names and treating one column as accession.
$ csvtk pretty -t example/taxonomy.tsv id superkingdom phylum class order family genus species --------------- ------------ -------------- ------------------- ---------------- ------------------ -------------- -------------------------- GCF_001027105.1 Bacteria Firmicutes Bacilli Bacillales Staphylococcaceae Staphylococcus Staphylococcus aureus GCF_001096185.1 Bacteria Firmicutes Bacilli Lactobacillales Streptococcaceae Streptococcus Streptococcus pneumoniae GCF_001544255.1 Bacteria Firmicutes Bacilli Lactobacillales Enterococcaceae Enterococcus Enterococcus faecium GCF_002949675.1 Bacteria Proteobacteria Gammaproteobacteria Enterobacterales Enterobacteriaceae Shigella Shigella dysenteriae GCF_002950215.1 Bacteria Proteobacteria Gammaproteobacteria Enterobacterales Enterobacteriaceae Shigella Shigella flexneri GCF_006742205.1 Bacteria Firmicutes Bacilli Bacillales Staphylococcaceae Staphylococcus Staphylococcus epidermidis GCF_000006945.2 Bacteria Proteobacteria Gammaproteobacteria Enterobacterales Enterobacteriaceae Salmonella Salmonella enterica GCF_000017205.1 Bacteria Proteobacteria Gammaproteobacteria Pseudomonadales Pseudomonadaceae Pseudomonas Pseudomonas aeruginosa GCF_003697165.2 Bacteria Proteobacteria Gammaproteobacteria Enterobacterales Enterobacteriaceae Escherichia Escherichia coli GCF_009759685.1 Bacteria Proteobacteria Gammaproteobacteria Moraxellales Moraxellaceae Acinetobacter Acinetobacter baumannii GCF_000148585.2 Bacteria Firmicutes Bacilli Lactobacillales Streptococcaceae Streptococcus Streptococcus mitis GCF_000392875.1 Bacteria Firmicutes Bacilli Lactobacillales Enterococcaceae Enterococcus Enterococcus faecalis GCF_000742135.1 Bacteria Proteobacteria Gammaproteobacteria Enterobacterales Enterobacteriaceae Klebsiella Klebsiella pneumonia # the first column as accession $ taxonkit create-taxdump -A 1 example/taxonomy.tsv -O example/taxdump 16:31:31.828 [INFO] I will use the first row of input as rank names 16:31:31.843 [INFO] 13 records saved to example/taxdump/taxid.map 16:31:31.843 [INFO] 39 records saved to example/taxdump/nodes.dmp 16:31:31.843 [INFO] 39 records saved to example/taxdump/names.dmp 16:31:31.843 [INFO] 0 records saved to example/taxdump/merged.dmp 16:31:31.843 [INFO] 0 records saved to example/taxdump/delnodes.dmp $ export TAXONKIT_DB=example/taxdump $ taxonkit list --ids 1 | taxonkit filter -E species | taxonkit lineage -r | csvtk pretty -Ht 793223984 Bacteria;Proteobacteria;Gammaproteobacteria;Moraxellales;Moraxellaceae;Acinetobacter;Acinetobacter baumannii species 1220345221 Bacteria;Proteobacteria;Gammaproteobacteria;Pseudomonadales;Pseudomonadaceae;Pseudomonas;Pseudomonas aeruginosa species 561101225 Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacterales;Enterobacteriaceae;Shigella;Shigella flexneri species 1969112428 Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacterales;Enterobacteriaceae;Shigella;Shigella dysenteriae species 599451526 Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacterales;Enterobacteriaceae;Escherichia;Escherichia coli species 2034984046 Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacterales;Enterobacteriaceae;Salmonella;Salmonella enterica species 1859674812 Bacteria;Proteobacteria;Gammaproteobacteria;Enterobacterales;Enterobacteriaceae;Klebsiella;Klebsiella pneumoniae species 773201972 Bacteria;Firmicutes;Bacilli;Bacillales;Staphylococcaceae;Staphylococcus;Staphylococcus aureus species 1295317147 Bacteria;Firmicutes;Bacilli;Bacillales;Staphylococcaceae;Staphylococcus;Staphylococcus epidermidis species 182402976 Bacteria;Firmicutes;Bacilli;Lactobacillales;Enterococcaceae;Enterococcus;Enterococcus faecium species 1566113429 Bacteria;Firmicutes;Bacilli;Lactobacillales;Enterococcaceae;Enterococcus;Enterococcus faecalis species 891083107 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;Streptococcus pneumoniae species 1357145446 Bacteria;Firmicutes;Bacilli;Lactobacillales;Streptococcaceae;Streptococcus;Streptococcus mitis species $ head -n 3 example/taxdump/taxid.map GCF_001027105.1 773201972 GCF_001096185.1 891083107 GCF_001544255.1 182402976 -
Custom lineages with the first row as rank names (pure lineage data)
$ csvtk cut -t -f 2- example/taxonomy.tsv | head -n 2 | csvtk pretty -t superkingdom phylum class order family genus species ------------ ---------- ------- ---------- ----------------- -------------- --------------------- Bacteria Firmicutes Bacilli Bacillales Staphylococcaceae Staphylococcus Staphylococcus aureus $ csvtk cut -t -f 2- example/taxonomy.tsv \ | taxonkit create-taxdump -O example/taxdump2 16:53:08.604 [INFO] I will use the first row of input as rank names 16:53:08.614 [INFO] 39 records saved to example/taxdump2/nodes.dmp 16:53:08.614 [INFO] 39 records saved to example/taxdump2/names.dmp 16:53:08.614 [INFO] 0 records saved to example/taxdump2/merged.dmp 16:53:08.615 [INFO] 0 records saved to example/taxdump2/delnodes.dmp $ export TAXONKIT_DB=example/taxdump2 $ taxonkit list --ids 1 | taxonkit filter -E species | taxonkit lineage -r | head -n 2 793223984 Bacteria;Proteobacteria;Gammaproteobacteria;Moraxellales;Moraxellaceae;Acinetobacter;Acinetobacter baumannii species 1220345221 Bacteria;Proteobacteria;Gammaproteobacteria;Pseudomonadales;Pseudomonadaceae;Pseudomonas;Pseudomonas aeruginosa species
genautocompleteLink
Usage
Generate shell autocompletion script
Supported shell: bash|zsh|fish|powershell
Bash:
# generate completion shell
taxonkit genautocomplete --shell bash
# configure if never did.
# install bash-completion if the "complete" command is not found.
echo "for bcfile in ~/.bash_completion.d/* ; do source \$bcfile; done" >> ~/.bash_completion
echo "source ~/.bash_completion" >> ~/.bashrc
Zsh:
# generate completion shell
taxonkit genautocomplete --shell zsh --file ~/.zfunc/_taxonkit
# configure if never did
echo 'fpath=( ~/.zfunc "${fpath[@]}" )' >> ~/.zshrc
echo "autoload -U compinit; compinit" >> ~/.zshrc
fish:
taxonkit genautocomplete --shell fish --file ~/.config/fish/completions/taxonkit.fish
Usage:
taxonkit genautocomplete [flags]
Flags:
--file string autocompletion file (default "/home/shenwei/.bash_completion.d/taxonkit.sh")
-h, --help help for genautocomplete
--type string autocompletion type (currently only bash supported) (default "bash")
profile2camiLink
Usage
Convert metagenomic profile table to CAMI format
Input format:
1. The input file should be tab-delimited
2. At least two columns needed:
a) TaxId of a taxon.
b) Abundance (could be percentage, automatically detected or use -p/--percentage).
Attention:
0. If some TaxIds are parents of others, please switch on -S/--no-sum-up to disable
summing up abundances.
1. Some TaxIds may be merged to another ones in current taxonomy version,
the abundances will be summed up.
2. Some TaxIds may be deleted in current taxonomy version,
the abundances can be optionally recomputed with the flag -R/--recompute-abd.
Usage:
taxonkit profile2cami [flags]
Flags:
-a, --abundance-field int field index of abundance. input data should be tab-separated (default 2)
-h, --help help for profile2cami
-0, --keep-zero keep taxons with abundance of zero
-S, --no-sum-up do not sum up abundance from child to parent TaxIds
-p, --percentage abundance is in percentage
-R, --recompute-abd recompute abundance if some TaxIds are deleted in current taxonomy version
-s, --sample-id string sample ID in result file
-r, --show-rank strings only show TaxIds and names of these ranks (default
[superkingdom,phylum,class,order,family,genus,species,strain])
-i, --taxid-field int field index of taxid. input data should be tab-separated (default 1)
-t, --taxonomy-id string taxonomy ID in result file
Examples
-
Test data, note that
2824115is merged to483329and1657696is deleted in current taxonomy version.$ cat example/abundance.tsv 2824115 0.2 merged to 483329 483329 0.2 absord 2824115 239935 0.5 no change 1657696 0.1 deleted -
Example:
$ taxonkit profile2cami -s sample1 -t 2021-10-01 \ example/abundance.tsv 13:17:40.552 [WARN] taxid is deleted in current taxonomy version: 1657696 13:17:40.552 [WARN] you may recomputed abundance with the flag -R/--recompute-abd @SampleID:sample1 @Version:0.10.0 @Ranks:superkingdom|phylum|class|order|family|genus|species|strain @TaxonomyID:2021-10-01 @@TAXID RANK TAXPATH TAXPATHSN PERCENTAGE 2 superkingdom 2 Bacteria 50.000000000000000 2759 superkingdom 2759 Eukaryota 40.000000000000000 74201 phylum 2|74201 Bacteria|Verrucomicrobia 50.000000000000000 6656 phylum 2759|6656 Eukaryota|Arthropoda 40.000000000000000 203494 class 2|74201|203494 Bacteria|Verrucomicrobia|Verrucomicrobiae 50.000000000000000 50557 class 2759|6656|50557 Eukaryota|Arthropoda|Insecta 40.000000000000000 48461 order 2|74201|203494|48461 Bacteria|Verrucomicrobia|Verrucomicrobiae|Verrucomicrobiales 50.000000000000000 7041 order 2759|6656|50557|7041 Eukaryota|Arthropoda|Insecta|Coleoptera 40.000000000000000 1647988 family 2|74201|203494|48461|1647988 Bacteria|Verrucomicrobia|Verrucomicrobiae|Verrucomicrobiales|Akkermansiaceae 50.000000000000000 57514 family 2759|6656|50557|7041|57514 Eukaryota|Arthropoda|Insecta|Coleoptera|Silphidae 40.000000000000000 239934 genus 2|74201|203494|48461|1647988|239934 Bacteria|Verrucomicrobia|Verrucomicrobiae|Verrucomicrobiales|Akkermansiaceae|Akkermansia 50.000000000000000 57515 genus 2759|6656|50557|7041|57514|57515 Eukaryota|Arthropoda|Insecta|Coleoptera|Silphidae|Nicrophorus 40.000000000000000 239935 species 2|74201|203494|48461|1647988|239934|239935 Bacteria|Verrucomicrobia|Verrucomicrobiae|Verrucomicrobiales|Akkermansiaceae|Akkermansia|Akkermansia muciniphila 50.000000000000000 483329 species 2759|6656|50557|7041|57514|57515|483329 Eukaryota|Arthropoda|Insecta|Coleoptera|Silphidae|Nicrophorus|Nicrophorus carolina 40.000000000000000 -
Recompute (normalize) the abundance
$ taxonkit profile2cami -s sample1 -t 2021-10-01 \ example/abundance.tsv --recompute-abd 13:19:23.647 [WARN] taxid is deleted in current taxonomy version: 1657696 @SampleID:sample1 @Version:0.10.0 @Ranks:superkingdom|phylum|class|order|family|genus|species|strain @TaxonomyID:2021-10-01 @@TAXID RANK TAXPATH TAXPATHSN PERCENTAGE 2 superkingdom 2 Bacteria 55.555555555555557 2759 superkingdom 2759 Eukaryota 44.444444444444450 74201 phylum 2|74201 Bacteria|Verrucomicrobia 55.555555555555557 6656 phylum 2759|6656 Eukaryota|Arthropoda 44.444444444444450 203494 class 2|74201|203494 Bacteria|Verrucomicrobia|Verrucomicrobiae 55.555555555555557 50557 class 2759|6656|50557 Eukaryota|Arthropoda|Insecta 44.444444444444450 48461 order 2|74201|203494|48461 Bacteria|Verrucomicrobia|Verrucomicrobiae|Verrucomicrobiales 55.555555555555557 7041 order 2759|6656|50557|7041 Eukaryota|Arthropoda|Insecta|Coleoptera 44.444444444444450 1647988 family 2|74201|203494|48461|1647988 Bacteria|Verrucomicrobia|Verrucomicrobiae|Verrucomicrobiales|Akkermansiaceae 55.555555555555557 57514 family 2759|6656|50557|7041|57514 Eukaryota|Arthropoda|Insecta|Coleoptera|Silphidae 44.444444444444450 239934 genus 2|74201|203494|48461|1647988|239934 Bacteria|Verrucomicrobia|Verrucomicrobiae|Verrucomicrobiales|Akkermansiaceae|Akkermansia 55.555555555555557 57515 genus 2759|6656|50557|7041|57514|57515 Eukaryota|Arthropoda|Insecta|Coleoptera|Silphidae|Nicrophorus 44.444444444444450 239935 species 2|74201|203494|48461|1647988|239934|239935 Bacteria|Verrucomicrobia|Verrucomicrobiae|Verrucomicrobiales|Akkermansiaceae|Akkermansia|Akkermansia muciniphila 55.555555555555557 483329 species 2759|6656|50557|7041|57514|57515|483329 Eukaryota|Arthropoda|Insecta|Coleoptera|Silphidae|Nicrophorus|Nicrophorus carolina 44.444444444444450 -
Some abundance might have taxa where some of them are parrents of others. E.g.,
$ cat example/abundance2.tsv 2 0.99 1224 0.59 1236 0.2 28211 0.4 1239 0.4 91061 0.39 2759 0.01 9606 0.01Please switch on -S/--no-sum-up to disable summing up abundances.
$ taxonkit profile2cami example/abundance2.tsv -S @SampleID: @Version:0.10.0 @Ranks:superkingdom|phylum|class|order|family|genus|species|strain @TaxonomyID: @@TAXID RANK TAXPATH TAXPATHSN PERCENTAGE 2 superkingdom 2 Bacteria 99.000000000000000 2759 superkingdom 2759 Eukaryota 1.000000000000000 1224 phylum 2|1224 Bacteria|Pseudomonadota 59.000000000000000 1239 phylum 2|1239 Bacteria|Bacillota 40.000000000000000 7711 phylum 2759|7711 Eukaryota|Chordata 1.000000000000000 28211 class 2|1224|28211 Bacteria|Pseudomonadota|Alphaproteobacteria 40.000000000000000 91061 class 2|1239|91061 Bacteria|Bacillota|Bacilli 39.000000000000000 1236 class 2|1224|1236 Bacteria|Pseudomonadota|Gammaproteobacteria 20.000000000000000 40674 class 2759|7711|40674 Eukaryota|Chordata|Mammalia 1.000000000000000 9443 order 2759|7711|40674|9443 Eukaryota|Chordata|Mammalia|Primates 1.000000000000000 9604 family 2759|7711|40674|9443|9604 Eukaryota|Chordata|Mammalia|Primates|Hominidae 1.000000000000000 9605 genus 2759|7711|40674|9443|9604|9605 Eukaryota|Chordata|Mammalia|Primates|Hominidae|Homo 1.000000000000000 9606 species 2759|7711|40674|9443|9604|9605|9606 Eukaryota|Chordata|Mammalia|Primates|Hominidae|Homo|Homo sapiens 1.000000000000000 -
Also see https://github.com/shenwei356/sun2021-cami-profiles
cami-filterLink
Usage
Remove taxa of given TaxIds and their descendants in CAMI metagenomic profile
Input format:
The CAMI (Taxonomic) Profiling Output Format
- https://github.com/CAMI-challenge/contest_information/blob/master/file_formats/CAMI_TP_specification.mkd
- One file with mutiple samples is also supported.
How to:
- No extra taxonomy data needed, so the original taxonomic information are
used and not changed.
- A mini taxonomic tree is built from records with abundance greater than
zero, and only leaves are retained for later use. The rank of leaves may
be "strain", "species", or "no rank".
- Relative abundances (in percentage) are recomputed for all leaves
(reference genome).
- A new taxonomic tree is built from these leaves, and abundances are
cumulatively added up from leaves to the root.
Examples:
1. Remove Archaea, Bacteria, and EukaryoteS, only keep Viruses:
taxonkit cami-filter -t 2,2157,2759 test.profile -o test.filter.profile
2. Remove Viruses:
taxonkit cami-filter -t 10239 test.profile -o test.filter.profile
Usage:
taxonkit cami-filter [flags]
Flags:
--field-percentage int field index of PERCENTAGE (default 5)
--field-rank int field index of taxid (default 2)
--field-taxid int field index of taxid (default 1)
--field-taxpath int field index of TAXPATH (default 3)
--field-taxpathsn int field index of TAXPATHSN (default 4)
-h, --help help for cami-filter
--leaf-ranks strings only consider leaves at these ranks (default [species,strain,no rank])
--show-rank strings only show TaxIds and names of these ranks (default
[superkingdom,phylum,class,order,family,genus,species,strain])
--taxid-sep string separator of taxid in TAXPATH and TAXPATHSN (default "|")
-t, --taxids strings the parent taxid(s) to filter out
-f, --taxids-file strings file(s) for the parent taxid(s) to filter out, one taxid per line
Examples:
- Remove Eukaryota
taxonkit profile2cami -s sample1 -t 2021-10-01 \ example/abundance.tsv --recompute-abd \ | taxonkit cami-filter -t 2759 @SampleID:sample1 @Version:0.10.0 @Ranks:superkingdom|phylum|class|order|family|genus|species|strain @TaxonomyID:2021-10-01 @@TAXID RANK TAXPATH TAXPATHSN PERCENTAGE 2 superkingdom 2 Bacteria 100.000000000000000 74201 phylum 2|74201 Bacteria|Verrucomicrobia 100.000000000000000 203494 class 2|74201|203494 Bacteria|Verrucomicrobia|Verrucomicrobiae 100.000000000000000 48461 order 2|74201|203494|48461 Bacteria|Verrucomicrobia|Verrucomicrobiae|Verrucomicrobiales 100.000000000000000 1647988 family 2|74201|203494|48461|1647988 Bacteria|Verrucomicrobia|Verrucomicrobiae|Verrucomicrobiales|Akkermansiaceae 100.000000000000000 239934 genus 2|74201|203494|48461|1647988|239934 Bacteria|Verrucomicrobia|Verrucomicrobiae|Verrucomicrobiales|Akkermansiaceae|Akkermansia 100.000000000000000 239935 species 2|74201|203494|48461|1647988|239934|239935 Bacteria|Verrucomicrobia|Verrucomicrobiae|Verrucomicrobiales|Akkermansiaceae|Akkermansia|Akkermansia muciniphila 100.000000000000000